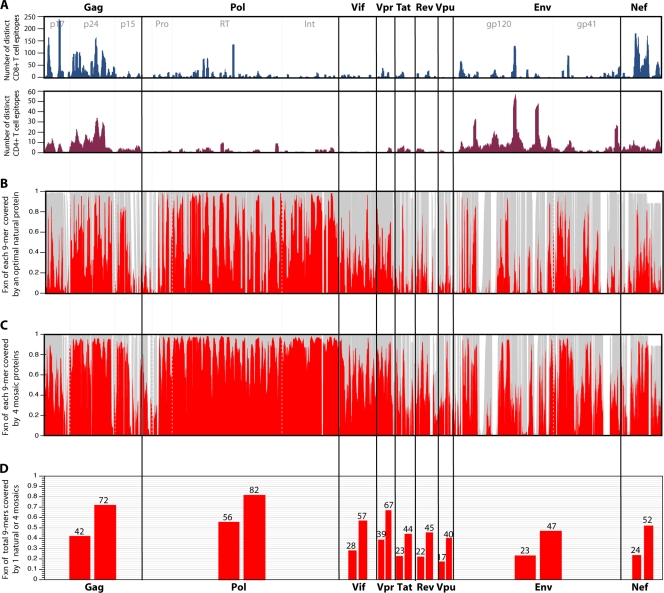
FIG. 3.
(A) Map of known and distinctive CD4+ and CD8+ epitopes described in the literature and included in the Los Alamos database across the proteome. Experimental maps of population responses (30) are tracked, with the exception of the underrepresentation of T-cell epitope mapping of regulatory proteins in the database. This underrepresentation is a consequence of regulatory protein T-cell epitope mapping being somewhat underrepresented in the literature. (B) The fraction (Fxn) of identical matches (red) with a single natural strain that provides the optimal coverage of the M group for each 9-mer (potential epitope) in the HIV proteome is shown. The optimal natural strain is a C-subtype sequence, C.ZA.99.DU422 (GenBank accession number AY043175). This is not surprising, as C is the most common subtype in the full-length genome database. This is an alignment-based figure; the gray background illustrates how many sequences have 9-mer in a given position in the alignment, such that a section in the alignment with an insertion in only one or a few sequences will appear as a white band. (C) Illustration of the increase in the fraction of perfectly matched 9-mers at each position when a four-mosaic combination is used rather than a single natural strain. (D) Total percentage of all 9-mers covered for each protein, corresponding to the single natural strain coverage shown in panel B (lower bar for each protein) and the four-mosaic coverage (higher bar) shown in panel C. The mosaic sequences were derived, optimal natural proteins were selected, and coverage graphics for both Fig. 3 and 4 were created using the mosaic vaccine tool suite at the Los Alamos HIV database (83) (http://www.hiv.lanl.gov/content/sequence/MOSAIC/). All comparisons were made to proteins translated from the global full-length genome alignment at the Los Alamos HIV database (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html), so the same input set of full genome sequences were used for every protein; thus, the variability comparisons between proteins are reasonable.