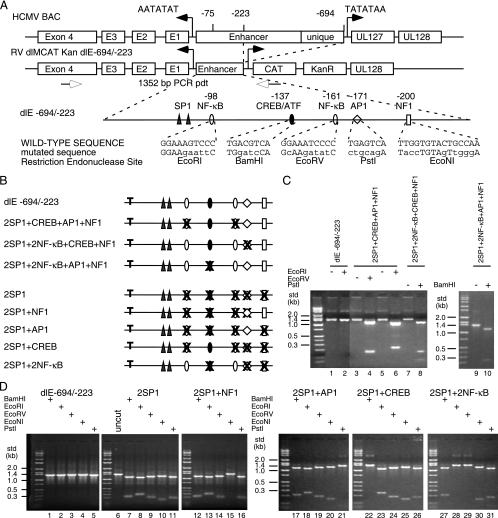
FIG. 2.
Construction of HCMV BAC mutations in the MIE proximal enhancer. (A) Diagram of the MIE enhancer/unique region flanked by the IE1 (UL123) and UL127 promoters. The distal enhancer and unique region were deleted between −694 and −223 relative to the IE1 transcription start site (+1) to isolate the proximal enhancer to −223. The UL127 open reading frame/modulator region was substituted by the CAT and kanamycin resistance (KanR) genes to generate RV dlMCAT Kan dl-694/-223 (dlE-694/-223). The known eukaryotic transcription-factor binding sites between −39 and −223 are designated, along with the mutated sequences and the newly generated restriction endonuclease sites. Each type of transcription factor binding site is designated by a symbol. (B) Summary diagram of mutations in the proximal enhancer with either one transcription factor binding site missing upstream of the two SP1 sites or one transcription factor binding site present. “T” designates the TATA box. The symbols for each type of transcription factor binding site are defined in panel A. (C and D) After PCR to generate the 1,352-bp DNA product (pdt) of the wild type or the recombinant HCMV DNAs with various mutations in the proximal enhancer region, the DNAs were digested with restriction endonucleases to prove an altered transcription factor binding site between −223 and −39. (D) HCMV BAC mutations with one transcription factor binding site present upstream of the two SP1 sites were constructed in both Towne BAC and FIX BAC. std, size standard.