Abstract
The DNA polymerase encoded by gene 5 (gp5) of bacteriophage T7 has low processivity, dissociating after the incorporation of a few nucleotides. Upon binding to its processivity factor, Escherichia coli thioredoxin (Trx), the processivity is increased to approximately 800 nucleotides per binding event. Several interactions between gp5/Trx and DNA are required for processive DNA synthesis. A basic region in T7 DNA polymerase (residues K587, K589, R590, and R591) is located in proximity to the 5′ overhang of the template strand. Replacement of these residues with asparagines results in a threefold reduction of the polymerization activity on primed M13 single-stranded DNA. The altered gp5/Trx exhibits a 10-fold reduction in its ability to support growth of T7 phage lacking gene 5. However, T7 phages that grow at a similar rate provided with either wild-type or altered polymerase emerge. Most of the suppressor phages contain genetic changes in or around the coding region for gene 3, an endonuclease. Altered gene 3 proteins derived from suppressor strains show reduced catalytic activity and are inefficient in complementing growth of T7 phage lacking gene 3. Results from this study reveal that defects in processivity of DNA polymerase can be suppressed by reducing endonuclease activity.
The efficient replication of the chromosome of bacteriophage T7 is accomplished by a replisome consisting of relatively few components, thus making it an excellent model system for studies on DNA replication. Phage T7 encodes most of the essential proteins required for DNA replication including gene 5 DNA polymerase (gp5), gene 4 helicase-primase, and gene 2.5 single-stranded DNA (ssDNA) binding protein (SSB) (39). One protein, thioredoxin (Trx), is encoded by the host Escherichia coli and serves as the processivity factor for T7 DNA polymerase. The three-dimensional structures of all four proteins have been determined (10, 20, 21, 43). Within the replisome, the helicase domain located in the C-terminal half of the gene 4 protein unwinds the double-stranded DNA (dsDNA) to provide an ssDNA template for the continuous synthesis of the leading strand by the gp5/Trx complex. The single-stranded lagging strand extruded behind the helicase is protected by the gene 2.5 protein until the primase domain located in the N-terminal half of the gene 4 protein synthesizes primers at primase recognition sites to initiate lagging-strand synthesis. The T7 replisome, reconstituted from these four proteins, mediates the replication of duplex DNA in a reaction where leading and lagging strands are coordinated (29). Coordination is achieved by the formation of a replication loop of the lagging strand containing the nascent Okazaki fragment.
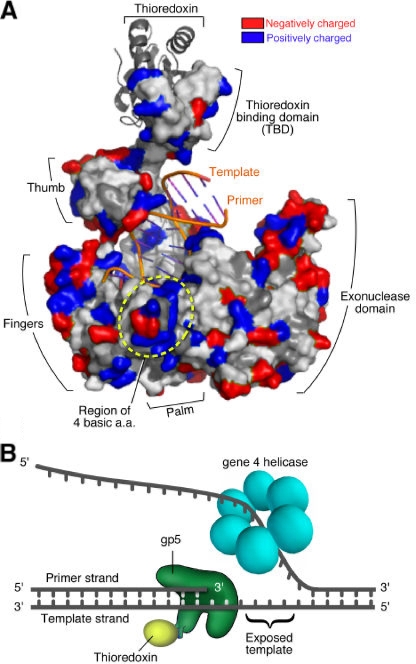
A key component of the T7 replisome is the gp5/Trx complex. Like most other prokaryotic DNA polymerases, it consists of a C-terminal polymerase domain and an N-terminal exonuclease domain (1, 28). As a member of the polymerase I family of DNA polymerases, it resembles a partially closed right hand with a thumb, palm, and fingers (Fig. 1A). The DNA binding within a crevice formed by the thumb, palm, and fingers leads to the catalytic site where the 3′-hydroxyl terminus of the primer strand is extended by the condensation of the properly positioned nucleoside triphosphate (NTP) in the nucleotide binding site. A distinguishing feature of T7 DNA polymerase is the presence of a unique segment of 76 residues at the tip of the thumb region; this segment is not present in other homologs of the polymerase I family. Trx binds to this region, designated the Trx-binding domain, with high affinity (25). The presence of Trx increases the processivity of the polymerase from the incorporation of 1 to 50 nucleotides to approximately 800 nucleotides per binding event (44). Two basic loops in the Trx-binding domain interact with the acidic C termini of both gene 4 and gene 2.5 proteins, suggesting that this region plays a regulatory role (18).
FIG. 1.
Interaction of T7 gp5/Trx complex with primer-template DNA. (A) Structure of primer-template-bound T7 gp5/Trx complex. A partially closed right-hand-shaped polymerase domain of gp5 (shown in surface model) in complex with E. coli Trx (gray ribbon model) (10). Trx is bound to a unique segment in the thumb subdomain of the polymerase. The primer-template (orange lines) resides in the DNA binding cleft created by the fingers, thumb, and palm. Colors in the polymerase represent electrostatic potential of residues exposed to the surface: red (acidic) and blue (basic). Four basic residues (K587, K589, R590, and R591) forming a positively charged patch nearby the 5′ end of the template strand are indicated. (B) Leading-strand DNA synthesis at replication fork. As the helicase domain of T7 gene 4 protein generates ssDNA template by unwinding parental dsDNA, gp5/Trx continuously synthesizes leading-strand DNA by extending primer in 5′ to 3′ direction. TBD, Trx-binding domain; aa, amino acid.
Several X-ray crystallographic structures of gp5/Trx in complex with a primer-template and a nucleoside 5′ triphosphate are available (2, 10, 12). The double-stranded portion of the primer-template in these structures is at the transition from B-form to A-form, implying significant distortions during catalysis (10). The DNA is held securely through numerous contacts with the polymerase where the 5′ end of the primer interacts with the thumb region and the 3′ end interacts with the fingers as well as the palm region (10). Such interactions facilitate alignment of the 3′-hydroxyl end of the primer to the active site of the enzyme where catalytically crucial residues reside. Further interactions with the incoming deoxy NTP (dNTP) induce conformational changes in the finger region to insure that only a properly base-paired dNTP can be incorporated (9). Thus, the structural environment around the active site is critical for efficient incorporation of the correct nucleotide.
Whereas the dsDNA region of the primer-template resides within a crevice formed by the thumb and fingers of the polymerase, the 5′ ssDNA overhang of the template strand lies outside the fingers. This region is less defined for interaction with the polymerase in the crystallographic structure. The 5′ nucleotides in the template strand adjacent to that hydrogen bonded with the incoming nucleotide are flipped out of the active site, and the last two 5′-terminal bases of the template are disordered (10). Clearly, however, this region of the template strand must be firmly anchored in order for the incoming nucleotide to hydrogen bond with the proper nucleotide in the template strand. The crystal structure of the polymerase reveals several patches of basic residues in proximity to the 5′ terminus of the template strand (10) (Fig. 1A). The possibility arises that these basic regions play a role in stabilizing the template for efficient DNA synthesis.
Other than the three essential proteins mentioned above, several additional T7-encoded proteins are likely to be involved directly or indirectly in DNA replication. For example, the T7-encoded 5′→3′ exonuclease (gp6) and DNA ligase (gp1.3) are involved in the removal of RNA primers from Okazaki fragments and the subsequent covalent joining of the fragments, respectively (14). In fact, approximately one-third of the T7 genes (class II; genes 1.4 to 6.3) may be dedicated to DNA metabolism (35). These genes include those essential for T7 replication (genes 2.5, 4, and 5), for the regulation of E. coli or phage transcription (genes 2 and 3.5), or for the degradation of DNA (genes 3 and 6). Among them, an endonuclease encoded by gene 3 exhibits multiple functions essential for phage growth. One of the primary roles played by T7 gene 3 endonuclease is the degradation of host chromosomal DNA for the eventual use of the nucleotide products for phage DNA synthesis (4). While the endonuclease hydrolyzes ssDNA more efficiently (approximately 150-fold) than dsDNA, it can still introduce nicks into dsDNA, eventually resulting in the conversion of the DNA into acid-soluble oligonucleotides (4). This nicking activity of gene 3 protein (gp3) plays the important role of resolving Holliday junctions that occur during homologous recombination and repair of T7 DNA (30).
A number of studies on the Holliday junction-resolving activity of the gene 3 endonuclease have revealed interactions of this enzyme with various DNA substrates. T7 endonuclease binds to four-way junction DNA as a homodimer with a high affinity (nanomolar range of dissociation constant), resulting in a 1,000-fold preference for a Holliday junction over dsDNA (8). Mutagenesis (37) and structural studies (16, 17) have shown a catalytic center that contains two metal binding sites surrounded by several charged residues, similar to that observed in type II restriction endonucleases (16). The protein is comprised of two domains, and exchanging the domains in dimeric conformation yields two composite active sites in which bilateral DNA cleavages occur (8). Recognition of the branched DNA appears to be initiated by the basic surface of the protein with structure-specific binding, which occurs through an induced-fit type of structural adjustment between DNA and the protein (17). Such a binding mode of the endonuclease suggests that the protein interacts with a broad range of DNA structures.
In the present study, we have examined the role of a basic cleft in T7 DNA polymerase that we considered likely to interact with the ssDNA region of the 5′ end of the bound template. Replacement of four basic residues in this cleft with uncharged asparagines results in a reduction of processive polymerase activity, whereas the exonuclease activity of the enzyme is not affected significantly. The altered polymerase is defective in supporting the growth of T7 phage lacking gene 5. We report that defects observed with the altered DNA polymerase can be suppressed by mutations in gene 3 endonuclease.
MATERIALS AND METHODS
Materials.
Oligonucleotides were obtained from Invitrogen or Integrated DNA Technology. Restriction endonucleases, alkaline phosphatase, M13 mp18 ssDNA, plasmid pUC(AT), and Deep Vent polymerase were from New England Biolabs. T4 polynucleotide kinase, T4 DNA ligase, radiolabeled nucleotides, and poly(dA) were purchased from Amersham Bioscience. Agarose was from USB Corp. DNA purification kits were from Qiagen.
Construction of plasmids, site-directed mutagenesis, protein overproduction, and purification.
Site-directed mutations were introduced into the T7 gene 5 contained in a plasmid (pGP5-3) following a standard PCR and cloning procedure (42). T7 DNA polymerases in complex with E. coli Trx was overproduced and purified as described previously (34). DNA encoding T7 gene 3 was inserted between the NdeI and HindIII sites of pET24a or pET28b plasmids (EMD Science), and site-directed mutations were introduced following a standard PCR and cloning procedure. The entire gene 3 coding region was confirmed by DNA sequence analysis. For overproduction and purification of His-tagged gp3s, pET28b derivatives containing gp3 (pET28-gp3) were transformed into E. coli HMS174(DE3) or HMS174(DE3)pLysS and grown to an optical density at 600 nm of 0.6 at room temperature. Genes were induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM, and the cells were grown for three additional hours. After harvest, cells were suspended in buffer A (20 mM Tris-HCl, pH 7.5, and 0.1 M NaCl) and ruptured by sonication. The cell lysate was collected by centrifugation and loaded onto a Ni-nitrilotriacetic acid affinity column. The Ni-nitrilotriacetic acid resin was washed with buffer A containing 50 mM imidazole, and gp3s were eluted with buffer A containing 500 mM imidazole (37). All of the purified proteins were greater than 90% pure as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie blue.
Construction of T7 deletion phage.
T7 phage lacking either gene 3 or both gene 3 and gene 5 were generated by homologous recombination as described previously (27). Overall, T7 genes to be deleted were replaced with the E. coli Trx-coding gene (trxA) in a plasmid, and then the plasmid was recombined with chromosomal DNA of T7 phage. Afterward, T7 gene deletion phage generated as the result of recombination were selected using Trx as a selection marker.
For construction of T7 phage lacking gene 3, plasmid pBRd3t was constructed by inserting a T7 DNA segment (position 9148 to 10926) into plasmid pBR322 between the EcoRI and PstI sites. A portion of the inserted DNA, part of gene 2.8, and the entire gene 3 (position 9880 to 10711) were replaced with trxA. Both pBRd3t and pET24-gp3 expressing gene 3 were transformed into E. coli A307 lacking Trx. After infection of the bacteria with wild-type T7 phage, the resulting lysate was collected and plated on E. coli A307 containing pET24-gp3 only. Since T7 phage requires Trx for DNA replication, only phage acquiring the Trx coding region from pBRd3t by recombination can grow on the E. coli A307. Recombinant phage plaques were isolated, and deletion of gene 3 was confirmed by PCR analysis.
For construction of T7 phage lacking both gene 3 and gene 5, the procedure described above was applied except that T7Δ5 lacking T7 gene 5 was used instead of wild-type T7 phage, and plasmid pGP5-3 was present to provide gene 5.
Construction of recombinant T7 phage containing suppressor mutations in gene 3.
To generate T7 phage containing mutations in specific positions of gene 3, mutations were introduced into gene 3, and then the altered gene 3 was introduced into T7 phage lacking both gene 3 and gene 5 by homologous recombination as described above. Plasmid pBR-supp was constructed by inserting a T7 suppressor DNA (position 9148 to 10926) between the EcoRI and PstI sites of pBR322. E. coli C600 containing pBR-supp and pGP5-3 was infected with the T7 phage lacking both gene 3 and gene 5. The lysate was collected and plated on E. coli strain C600 containing only pGP5-3 without a plasmid expressing gene 3. Acquisition of gene 3 in the generated phage was confirmed by PCR analysis. To rule out additional genetic alterations that could occur during recombination, the DNA sequence (positions 9148 to 11060) in the recombinant phage was verified by sequencing analysis.
Phage growth complementation assay.
The ability of T7 phage to grow on E. coli was determined by spot or plaque assay. First, to identify the range of phage concentration, an aliquot of an E. coli culture grown at log phase (optical density at 600 nm of ∼1) was mixed with soft agar and plated on an LB plate containing the appropriate antibiotics. A small amount (1 to 2 μl) of serially diluted phage solution was spotted and grown at 37°C for 5 to 6 h or at room temperature overnight. Once the concentration of phage stock was determined, 0.3 ml of E. coli culture grown at log phase was mixed with 0.1 ml of diluted phage stock, plated on an LB plate, and incubated as described above.
DNA polymerase assay for T7 gp5/Trx.
The standard T7 DNA polymerase assay contained either M13 ssDNA annealed to a 24-mer primer (20 nM) or poly(dA)390 annealed to poly(dT)25 (200 nM); 0.25 mM each of [α-3H]dTTP (5 cpm/pmol), dATP, dCTP, and dGTP; 40 mM Tris-HCl, pH 7.5; 10 mM MgCl2; 10 mM dithiothreitol (DTT); 50 mM potassium glutamate; and the amounts of T7 gp5/Trx indicated in the figures. Incubation was for 10 min at 37°C for the primed M13 template or 5 min at room temperature for the poly(dA)/oligo(dT). After the reaction was terminated by the addition of EDTA to a final concentration of 20 mM, aliquots were spotted onto DE-81 filters (Whatman) and washed three times with ammonium formate. The amount of nucleotide incorporated by gp5/Trx was determined by measuring the radioactivity retained on the filters.
For an analysis of the products obtained from DNA synthesis reactions using primed M13 ssDNA, the primer was radiolabeled at its 5′ end with 32P using polynucleotide kinase. After incubation at 37°C for 20 min, the reaction products were separated on 0.6% agarose gel containing ethidium bromide (0.3 μg/ml) and analyzed by autoradiography.
To examine the effect of E. coli SSB protein on polymerase activity, 20 nM primed M13 ssDNA; 4 nM T7 gp5/Trx; and 0.25 mM each of [α-3H] dTTP (5 cpm/pmol), dATP, dCTP, and dGTP were incubated in the presence or absence of 10 μM E. coli SSB protein at 37°C for the time period indicated in the figures. Incorporation of [3H]dTMP was determined as described above.
Exonuclease assay for T7 gp5/Trx.
Exonuclease activity of T7 DNA polymerase was measured using uniformly 3H-labeled ss- or dsDNA prepared as described previously (26). The assay contained either 2 nM dsDNA or 1.2 nM ssDNA, 40 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM DTT, 50 mM potassium glutamate, and the amounts of T7 gp5/Trx indicated in the figures. Incubation was at 37°C for 10 min. The radioactivity remaining in DNA after incubation was determined using a DE-81 filter as described above.
Endonuclease assay for T7 gene 3 endonuclease.
The standard assay for gene 3 endonuclease contained the indicated DNA (30 nM), 50 mM Tris-HCl, pH 7.9, 10 mM MgCl2, 1 mM DTT, 100 mM NaCl, and the amount of gene 3 endonuclease indicated in the figures. After incubation at 37°C for 20 min, the reaction products were separated on 1% agarose gel containing ethidium bromide. The amount of hydrolysis of the DNA was determined by measuring the intensity of DNA bands using an Alpha Imager3400.
RESULTS
As the DNA helicase bound to the lagging strand unwinds the duplex DNA at the replication fork, the newly created ssDNA template is copied by the DNA polymerase bound to the leading strand (Fig. 1B). Although the length of the ssDNA template at the replication fork is not known, it may be dictated by the spatial arrangement of the helicase and polymerase. It seems likely that this stretch of template strand is bound to the polymerase so as to stabilize the template for proper base-pairing of the incoming NTP. Several crystallographic structures of T7 gp5/Trx in complex with primer-template illustrate the interaction of the polymerase with DNA (2, 10, 12), particularly at the active site where the incoming dNTP is located. However, only a few nucleotides of the ssDNA template in these studies diffract, the remaining region probably being disordered. Inspection of the structure of T7 DNA polymerase reveals a patch of four basic residues (K587, K589, R590, and R591) located in a region that has the potential to interact with the 5′ end of the template (Fig. 1A). In order to investigate the role of this basic region on polymerase activity, we replaced these residues with asparagines having a neutral side chain and examined the effect of these changes on T7 growth and on DNA polymerase activity. The altered gp5 (gp5-4N) as well as the wild-type protein was purified as a complex with Trx and used for biochemical assay.
DNA polymerase activity of altered gp5/Trx.
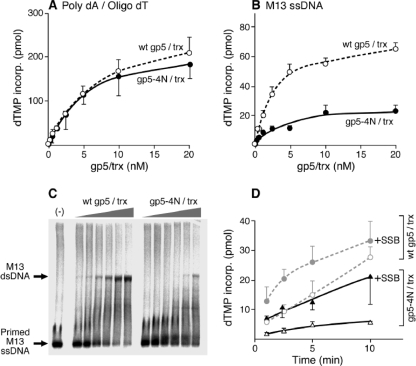
We first examined the ability of the altered DNA polymerase, gp5-4N/Trx, to incorporate deoxynucleoside monophosphate (dNMP) using poly(dA) template (average, 390-mer) to which an oligo(dT) primer of 25 nucleotides was annealed. The altered polymerase exhibits polymerase activity similar to that of wild-type T7 gp5/Trx (Fig. 2A). The results indicate that alteration at this region of the polymerase does not affect its catalytic activity as measured by its ability to add nucleotides to the 3′-OH of the primer strand. However, when primed M13 ssDNA was used in the assay, gp5-4N/Trx exhibited threefold lower activity than the wild-type protein (Fig. 2B). Analysis of the reaction products on agarose gel reveals that a low level of incorporation of dNMP by the altered polymerase is due to lower processivity than that found with the wild-type protein (Fig. 2C). The low processivity is reflected by the presence of primers not extended, the absence of fully replicated M13 dsDNA, and the presence of large amounts of aborted product of low mobility.
FIG. 2.
Polymerase activity of gp5-4N/Trx. (A) DNA synthesis on poly(dA)/oligo(dT) template. The reaction mixture described in Materials and Methods contained 200 nM poly(dA)390 annealed with oligo(dT)25; 0.25 mM each of 3H-labeled dTTP (5 cpm/pmol), dGTP, dATP, and dCTP; and the indicated amounts of gp5-4N/Trx or wild-type (wt) gp5/Trx. After incubation at room temperature for 5 min, the activity of DNA polymerase was determined by measuring the incorporation (incorp) of [3H]dTMP into DNA. (B) DNA synthesis on primed M13 ssDNA. The reaction mixture was similar to that described for panel A except that 20 nM M13 ssDNA annealed to a primer was used as a template instead of poly(dA)/oligo(dT). Reaction mixtures were incubated at 37°C for 10 min. (C) Products from DNA synthesis on primed M13 ssDNA template. Radioactively end-labeled 5′ 32P-labeled 24-mer primer was annealed to M13 ssDNA and incubated with increasing amounts of gp5/Trx (0.3, 0.5, 1, 2, 4, and 8 nM) at 37°C for 20 min. Reaction products were analyzed on 0.6% agarose gel and dried for autoradiography. Locations of the initial template and full-length product M13 dsDNA are indicated. (D) Effect of E. coli SSB protein on DNA synthesis by gp5/Trx. Reaction mixture contained 20 nM primed M13 ssDNA template, 4 nM gp5/Trx, and 0.25 mM all four dNTPs (5 cpm/pmol [3H]dTTP). The reaction mixture was incubated in the absence or presence of 10 μM E. coli SSB protein at 37°C for the indicated time period. Amount of incorporated [3H]dTMP into DNA was measured.
We have previously shown that secondary structures in M13 ssDNA can hamper progression of T7 gp5/Trx and that the addition of E. coli SSB protein can resolve the pausing of the polymerase (26). Indeed, addition of E. coli SSB protein enhanced incorporation of dNMP by both DNA polymerases (Fig. 2D). However, the altered polymerase still exhibited lower activity than the wild-type enzyme.
Exonuclease activity of altered gp5/Trx.
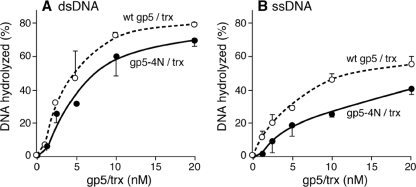
Like other DNA polymerases in the polymerase I family, T7 gp5 contains an exonuclease domain located in the N terminus of the protein. The exonuclease is active on both ssDNA and dsDNA, with Trx stimulating the latter (23). gp5-4N/Trx has somewhat less exonuclease activity on both ds- and ssDNA (80 and 60%, respectively) than the wild-type enzyme (Fig. 3). Again, the lower activity with gp5-4N/Trx may reflect its lower processivity, a parameter that affects exonuclease activity (25, 44).
FIG. 3.
Exonuclease activity of T7 DNA polymerase. Either 2 nM M13 3H-labeled dsDNA (A) or 1.2 nM M13 3H-labeled ssDNA (B) was incubated in the reaction mixture described in Materials and Methods with the indicated concentration of either wild-type (wt) gp5/Trx or gp5-4N/Trx at 37°C for 10 min. Exonuclease activity was determined by measuring the radioactive DNA that bound to a DE-81 filter.
Complementation for growth of T7 phage lacking gene 5.
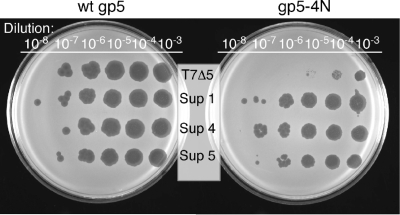
The effect of the alteration of T7 DNA polymerase on its in vivo function was examined by phage complementation assay. In this assay, growth of T7 phage lacking gene 5 is dependent on DNA polymerase exogenously expressed by a plasmid in the infected cell harboring functional gene 5. The replacement of the four basic residues with asparagines in gp5-4N results in a reduction of its ability to complement growth of the deletion phage by more than 10-fold (Fig. 4, right). The plaques produced by the infection are 10-fold smaller than those obtained when the wild-type protein is used for complementation (data not shown). Consistent with the biochemical data, the in vivo complementation assay indicates that the alteration severely reduces gp5 function.
FIG. 4.
In vivo complementation of phage by T7 gp5. Serial dilutions of either T7 gene 5-deletion phage or suppressor phages were spotted onto a LB plate containing E. coli expressing the indicated gp5. After incubation at 37°C overnight, the ability to lyse E. coli was compared. Sup, suppressor.
Isolation of suppressor phage and analysis of genetic changes.
From the above data, it is clear that the alteration of polymerase yields significant defects of in vivo function of the polymerase: reduction in both the ability to complement growth of the gene 5-deletion phage and the size of plaques. However, some plaques isolated from the culture with the altered polymerase yielded phage that produced larger plaque sizes upon replating on E. coli expressing gp5-4N. Those isolated suppressor phages grow with gp5-4N as efficiently as with the wild-type protein (Fig. 4).
To identify the genetic changes responsible for the gain of function in the complementation assay, we selected 10 suppressor phages from three independent trials. Since class II genes of the T7 genome (gene 1.4 to gene 6.3) are known to be involved in DNA metabolism, we determined the DNA sequence of DNA isolated from these suppressor phages. As summarized in Table 1, two suppressors out of a total of the 10 phages examined do not have any mutation in the class II genes. The remaining eight suppressors contain genetic alterations in or around gene 3. Three of the suppressors have single (suppressors 1 and 4) or double (suppressor 5) substitutions in gene 3, two of them have frameshifts in gene 2.8 (suppressors 6 and 7), immediately upstream of gene 3, and the remaining three contain deletions between gene 3.5 and 3.8 (suppressors 8, 9, and 10), downstream of gene 3. Five phages containing either frameshift or an intergenic deletion appear to originate from the same parental suppressor since they have the same sequence. Since the majority of suppressor phages have changes that may affect gene 3 function or expression, we chose the simplest alterations in gene 3, three substitutions (suppressors 1, 4, and 5) for further analysis.
TABLE 1.
Genetic alterations found in T7 suppressor phagesa
| Trial | Suppressor no. | Genetic alteration (nt)b | Outcome in gene expression |
|---|---|---|---|
| A | 1 | T10422C | gp3 F56L |
| B | 2 | ||
| 3 | |||
| C | 4 | A10482G | gp3 K76E |
| 5 | A10575G | gp3 T107A | |
| T10666C | gp3 V137A | ||
| 6 | Δ10132 | Frameshift in gene 2.8 | |
| 7 | Δ10132 | Frameshift in gene 2.8 | |
| 8 | Δ11195-11196 | Deletion in noncoding region between genes 3.5 and 3.8 | |
| 9 | Δ11195-11196 | Deletion in noncoding region between genes 3.5 and 3.8 | |
| 10 | Δ11195-11196 | Deletion in noncoding region between genes 3.5 and 3.8 |
A total 10 suppressor phages were isolated from three independent trials, and their DNA sequences in class II genes (gene 1.4 to gene 6.3) were analyzed. Genetic changes found from the phages and their consequences in gene expression are indicated.
nt, nucleotide.
Confirmation of gene 3 alteration in suppressor phage.
To establish that mutations in gene 3 are the sole determinant for suppression of the defect in gp5-4N, we constructed T7 phage lacking gene 5 but containing point mutations in gene 3 using homologous recombination. The recombinant phage contained single- or double-amino acid substitutions in gene 3 identified from the suppressor DNA sequences. These recombinant phages lacking gene 5 had similar growth when either wild-type gp5 or gp5-4N was provided from plasmids (data not shown). These results establish that mutations in gene 3 are responsible for the observed phenotype.
Complementation of phage lacking gene 3.
The effect of the mutations in gene 3, identified from suppressor phages, on phage growth was examined by a phage growth complementation assay. All of the mutations lead to gp3s that are defective in their ability (5- to 30-fold reduction in plating efficiency) to complement T7 phage lacking gene 3 (T7Δ3) (Table 2). The size of plaques produced by expression of the altered gp3s was also significantly smaller than the size produced by the wild-type protein (about threefold). We also examined the effect of alteration of two residues (E20 and D55) previously identified as critical factors for catalytic activity (8) on the ability to complement T7 phage lacking gene 3. Alteration of those residues (E20Q or D55N) also resulted in proteins with reduced ability to complement T7Δ3 as well as the production of small plaques (Table 2). However, these latter reductions in gene 3 function were more severe than those observed with the alterations identified in the suppressor analysis.
TABLE 2.
Complementation of T7 gene 3-deletion phage growth by recombinant proteinsa
| Protein expressedb | Alteration | Efficiency of platingc |
|---|---|---|
| None | 0.01* | |
| gp5 wt | wt | 6 × 10−4* |
| gp5-4N | K587N, K589N, R590N, R591N | 5 × 10−4* |
| gp3 wt | wt | 1.0 |
| gp3 (Supp 1) | F56L | 0.1* |
| gp3 (Supp 4) | K76E | 0.2* |
| gp3 (Supp 5) | T107A, V137A | 0.03* |
| gp3 (E20Q) | E20Q | 0.003* |
| gp3 (D55N) | D55N | 0.007* |
Recombinant proteins were expressed from plasmid under the control of a T7 promoter in E. coli C600. After infection with T7 gene 3-deletion phage (T7Δ3), the number of plaques were counted, normalized to the value obtained with wild-type gene 3, and results are presented as efficiency of plating. Data were obtained from at least duplicated experiments. E. coli cells expressing no protein or gp5 were used as negative controls. wt, wild-type; Supp, suppressor.
Alterations/mutations are given in parentheses.
*, small size of plaque.
Endonuclease activity of gp3s.
Gene 3 of bacteriophage T7 is essential for phage viability (5). Gene 3 encodes an endonuclease that is 100-fold more active on ssDNA than on dsDNA (4). A major role of the gene 3 endonuclease is, in concert with the gene 6 exonuclease, to degrade the host DNA so that the resulting nucleoside 5′ phosphates can be used for phage DNA synthesis (35). It also participates in homologous recombination by cleaving Holliday junctions (30). In order to examine the endonuclease activity of the gp3s arising from suppressor mutations, we purified them, along with the wild-type protein, as His-tagged recombinant proteins. The presence of the His tag on the N terminus of the endonuclease does not alter gp3 function, as judged by the ability of the modified gene to support growth of T7 phage lacking gene 3 (data not shown).
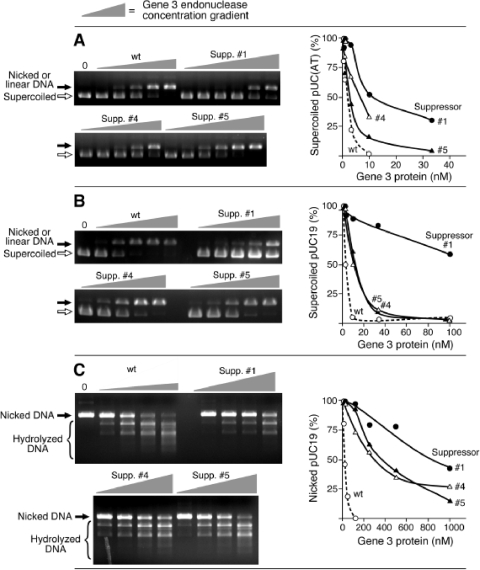
Gene 3 endonuclease hydrolyzes AT-rich regions with high efficiency (15). As shown in Fig. 5A, wild-type endonuclease rapidly hydrolyzes a supercoiled plasmid containing a stretch of alternating AT sequence (∼20 bp) to the linear or nicked form. All three of the endonucleases derived from gene 3 harboring suppressor mutations exhibited reduced activity, with the most severe defect (10-fold reduction) observed with suppressor 1. Nucleolytic digestion on a normal dsDNA plasmid, pUC19 (Fig. 5B), or nicked dsDNA plasmid pUC19 (Fig. 5C) requires more endonuclease to achieve a similar level of digestion (Table 3). Again, all altered proteins exhibit lower catalytic activity than that found with the wild-type protein (Fig. 5B and C). The most significant reduction of activity was observed with suppressor 1. The amount of each enzyme required to convert approximately 50% of the initial DNA into hydrolyzed forms is presented in Table 3. Catalytically inactive endonucleases, gp3 with the mutation E20Q (gp3-E20Q) and gp3-D55N, have severe reductions in activity, far more than enzymes derived from the suppressors.
FIG. 5.
Catalytic activity of gene 3 endonuclease. (A) Double-stranded plasmid pUC(AT) (30 nM) containing a sequence of approximately 20 bp of alternating AT sequence was incubated with increasing amounts of the indicated gp3s (0.1, 0.3, 1, 3, and 10 nM for wild type and suppressor 4; 0.1, 0.3, 1, 3, 10, and 30 nM for suppressors 1 and 5) at 37°C for 20 min. Reaction products were analyzed on 1% agarose gel. Initial supercoiled substrate and linear or nicked products are indicated on the left side of the gel. (B) Double-stranded plasmid pUC19 (30 nM) was incubated with increasing amounts of the indicated gp3s (1, 3, 10, 30, and 100 nM) at 37°C for 20 min. Reaction products were analyzed on 1% agarose gel. (C) Nicked plasmid was prepared by digesting pUC19 with endonuclease Nt.BstNBI. After inactivation of Nt.BstNBI by heating, the resulting nicked DNA (30 nM) was incubated with increasing amounts of gp3 (13, 25, 50, and 100 nM for the wild-type; 125, 250, 500, and 1,000 nM for the suppressor endonucleases) at 37°C for 20 min. Reaction products were analyzed on 1% agarose gel. Locations of initial substrate and cleavage product are indicated on the left side of the gel. wt, wild type; Supp, suppressor.
TABLE 3.
Nuclease activity of T7 gene 3 endonucleases
| Substrate | Activity of the indicated gene 3 endonuclease (nM)b
|
|||||
|---|---|---|---|---|---|---|
| wt | Supp 1 | supp 4 | Supp 5 | E20Q | D55N | |
| pUC(AT)a | 1 ± 0.2 | 12 ± 1 | 5 ± 1 | 2 ± 0.4 | 210 ± 26 | >1000 |
| pUC19 | 4 ± 1 | 116 ± 16 | 13 ± 3 | 16 ± 2 | ND | ND |
| Nicked pUC19 | 24 ± 7 | 775 ± 127 | 338 ± 59 | 369 ± 75 | ND | ND |
pUC(AT) contains a sequence of approximately 20 bp of alternating AT residues.
Nuclease reactions were carried out as described in the legend of Fig. 5. Numbers are nanomolar concentrations at which the indicated T7 gene 3 endonuclease converts approximately 50% of the initial DNA substrate into hydrolyzed forms. Data were obtained from at least duplicated experiments. wt, wild-type gp3; Supp, gp3 containing amino acid substitution derived from indicated suppressor phage; E20Q and D55N, gp3s containing the indicated single amino acid substitution; ND, not determined.
General effect of suppressor-derived gene 3 mutations on gp5/Trx having low processivity.
Is the ability of gene 3 endonuclease having reduced activity to suppress the phenotype of gp5-4N specific for the alterations in the basic patch of T7 gp5 or for low processivity of gp5/Trx in general? We have examined the ability of the gene 3 endonuclease mutants to suppress the defect in growth of T7 phage arising from another DNA polymerase of low processivity.
Gene 5 DNA polymerase normally forms a tight complex with its processivity factor E. coli Trx via a hydrophobic interaction (24). If E. coli Trx is covalently linked to gp5 through a disulfide bond (T327C in gp5 and C32 in Trx), the processivity is reduced (26). This DNA polymerase, in covalent complex with Trx, has a 100-fold reduction in its ability to complement the growth of T7 phage lacking gene 5 (26), considerably weaker complementation than the 10-fold reduction observed with gp5-4N (Table 4). However, suppressor phages lacking gene 5 but containing a defective gene 3 grow equally well when complemented with either wild-type gp5/Trx or the covalently linked gp5/Trx. These results suggest that the alterations of gene 3 contained in suppressors are beneficial to gp5s of low processivity.
TABLE 4.
Ability of gene 3 mutations to suppress T7 DNA polymerases of low processivity
| Phage | Complementation efficiency with the indicated mutant protein(s) [no. of PFU with mutated protein(s)/no. of PFU with wt protein(s)]a
|
|
|---|---|---|
| gp5-4N/gp5 wt | [gp5(T327C)/Trx(C35S)]/(gp5 wt/Trx wt) | |
| T7Δ5 | 0.08 | 0.006 |
| Suppressor 1 | 0.6 | 0.5 |
| Suppressor 4 | 0.9 | 0.2 |
| Suppressor 5 | 0.6 | 0.5 |
Recombinant proteins were expressed from plasmids under the control of a T7 promoter in either E. coli strain C600 (for single gene 5 expression) or E. coli A307 (for both gene 5 and Trx expression). After infection with the indicated T7 phage, the number of plaques was counted, and PFU were determined. Mutated residues are in parentheses. Data were obtained from at least two experiments. wt, wild-type protein.
DISCUSSION
Factors contributing to processive polymerization by T7 DNA polymerase.
Processive DNA polymerases maintain a stable binding to DNA in order to prevent their dissociation from the DNA after the condensation of each nucleotide. DNA polymerases of the polymerase I family resemble a partially closed right-handed structure containing a DNA binding crevice to accommodate primer-template. Numerous interactions within the crevice align the 3′-hydroxyl of the primer strand at the active site for incorporation of an incoming dNTP bound to the template strand. To achieve such stable binding to the DNA, most replicative DNA polymerases associate with processivity factors such as the β-sliding clamp or PCNA that encircles DNA (38). Topological confinement of the DNA with the ring-shaped processivity factor provides for efficient polymerization of nucleotides without dissociation from the DNA. T7 DNA polymerase is unique in that its processivity factor, E. coli Trx, binds to the polymerase with nanomolar binding affinity (24). However, it does not appear to encircle DNA, and the crystal structure of the complex reveals that it positions over the duplex region of the DNA (10). Disruptions of the association between the polymerase and Trx by altering either of the proteins leads to significant loss in processivity of the polymerizing complex (19, 24, 26, 46).
Stable contacts of the polymerase with the primer-template within the DNA binding crevice and in the pocket containing the active site are also likely to be another major contributor to processivity. For example, the occupancy of the nucleotide binding site by the properly hydrogen-bonded incoming NTP should be provided for considerable stability of the complex. At this point in the polymerization cycle, the fingers of the polymerase have rotated inward 41° so that several residues grasp the NTP prior to condensation and position it for nucleophilic attack by the 3′ hydroxyl of the primer (10). We have now shown that a basic patch in the polymerase is also important in processivity. This basic patch is in close proximity to the template strand as it exits the nucleotide binding site. Elimination of the positive charge of this patch by replacing the basic residues with asparagines leads to a significant decrease in processivity without affecting the other catalytic activities of the enzyme. It is reasonable to postulate that the elimination of charge results in a loss of contact of the negatively charged DNA with this patch and thus accounts for the loss in processivity. Unfortunately, at present there is no definitive way to examine specific binding of the template strand to the polymerase as it exits from the active site other than a crystal structure of the wild-type and altered polymerase in which the exiting template strand diffracts. Thus far, this has not been possible, perhaps due to the length of the primer-template used for crystallization. The duplex region of the primer-template binds within the DNA binding crevice of T7 DNA polymerase and is secured by its processivity factor Trx. Consequently, it is likely that this much tighter binding will mask any binding by the ssDNA template strand. However, we did examine the binding affinity of DNA polymerase to short DNA (either 65-mer ssDNA or 65-mer template annealed to a 22-mer primer) using a gel mobility shift assay. It seemed possible that binding of ssDNA alone to this basic patch could be detected in the absence of the primer-template structure. Indeed, gp5-4N binds to ssDNA slightly less than does wild-type polymerase (data not shown). As expected, no difference was observed between polymerases in binding to primer-template composed of the 65-mer annealed to a 22-mer primer.
Structural impedance such as chemically induced lesions or secondary structure in the template leads to dissociation of the polymerase and thus lower processivity (12, 36, 45). The structural hindrance can, in some cases, be bypassed to produce newly synthesized DNA lacking a few nucleotides found in the template. In other instances, the polymerase may continue by incorporating an incorrectly base-paired nucleotide (2). Addition of SSB proteins such as E. coli SSB protein can resolve the secondary structure (26). From the above, it is clear that the processivity of a polymerase is the result of at least two parameters. The structure of the DNA itself, as just discussed, provides one parameter. Another is the affinity of the polymerase for the primer-template. The affinity of most replicative DNA polymerases is significantly increased by processivity factors that by themselves bind tightly to DNA or induce conformation changes in the polymerase to increase the affinity for DNA. The low processivity of the DNA polymerase examined in this study appears to arise from a decreased affinity of the polymerase for the DNA. The finding that SSB protein does not overcome the low processivity supports this conclusion.
Molecular basis of suppression of polymerase defect by reduced gene 3 endonuclease activity.
Our results suggest that T7 phage can overcome defects in the processivity of DNA polymerase by suppressing the activity of gene 3 endonuclease. Among the T7 class II genes examined in our identification of suppressors, three out of six suppressors contained alterations exclusively in gene 3. All of the mutations reduced gene 3 function in vivo and reduced the endonuclease activity of the protein in vitro. The altered residues are either very close to residues critical to catalysis (F56L) or interact directly with the Holliday junction (K76E and T107A) (16, 17). Among the other suppressors not located in gene 3, two contain a deletion either immediately upstream (gene 2.8) or downstream (between gene 3.5 and 3.8) of gene 3. It has been proposed that alterations in the gene 2.8 region affect the translation of gp3 since the ribosomal binding site for gene 3 overlaps with the gene 2.8 coding region (40). Similarly, changes in the downstream intergenic region could modify the translation level of gp3 since it contains a potential RNase III cleavage site (11). Therefore, it is reasonable to speculate that the alterations identified near gene 3 influence the expression of gene 3, thereby reducing the level of endonuclease. Nevertheless, the fact that two suppressors do not contain any genetic alterations in the class II genes indicates another pathway can also overcome the defect in DNA polymerase.
How do mutations in gene 3 suppress the defects arising from mutations in the basic patch of T7 DNA polymerase? The most obvious explanation centers around the function of gene 3 and the biochemical defect in gp5-4N. Gene 3 encodes an endonuclease, and, indeed, all of the suppressor mutations decrease the endonuclease activity. gp5-4N/Trx is defective in processive DNA synthesis, and it is logical to suggest that the lower endonuclease activity in phage-infected cells somehow restores processivity. However, it is difficult to envision a mechanism by which an endonuclease with any level of activity could interact with a DNA template in vitro to increase processivity. We consider that it is more likely that the low processivity of the polymerase gives rise to DNA structures in the template that are prone to be cleaved by the gene 3 endonuclease. Such cleavage of the template strand could account for the poor growth of T7 phage relying upon gp5-4N for DNA replication. As described earlier, a DNA polymerase of low processivity has difficulty in copying a template containing secondary structure, often pausing and eventually dissociating during DNA synthesis. The paused replicating complex can be regarded as a stalled replication fork, a vulnerable target for the endonuclease activity of gene 3 protein (22). An endonuclease action on the stalled fork generated by a DNA polymerase of low processivity would be detrimental to DNA replication. Therefore, an endonuclease having reduced activity might not cleave at the stalled complex. Although this protective effect of the reduced endonuclease activity on DNA polymerization by gp5-4N/Trx was observed, our attempts to detect enhanced DNA synthesis in the presence of endonuclease were unsuccessful (data not shown). Additional proteins may be required to rescue low-processive polymerase.
It is noteworthy that all of the alterations in gp3 derived from suppressors retain their catalytic activity, which is not a surprising result since gene 3 endonuclease is important in degrading the host DNA in order to supply the T7 replication system with nucleotides (35) and in recombination in phage-infected cells (30). A replication fork whose progression is blocked due to damage of the DNA can be reactivated through recombination, replication restart, and resolution of the resulting DNA products (7). Recombination and repair proteins recognize and cleave Holliday junctions formed at stalled replication forks (32). Therefore, junction-resolving enzymes such as T7 gene 3 endonuclease that specifically recognize and cleave aberrant DNA structures are crucial for recombination-dependent replication (8). Although it is not yet demonstrated in the T7 replication system, it is likely that gene 3 endonuclease, known to recognize a variety of aberrant structures and to cleave Holliday junctions, plays a similar role. Thus, DNA polymerases of low processivity could be rescued through a recombination-mediated mechanism. For example, DNA synthesis at a stalled complex could be reinitiated by interaction with other recombination components such as T7 gene 2.5 ssDNA binding protein or gene 4 helicase-primase.
Compensatory evolution of T7 phage.
The self-sufficient genome organization of bacteriophage T7 and its robust lytic life cycle have made it an attractive model to investigate genetic adaptation under environmental pressure (13). One study has shown that T7 phage modifies expression of several genes in the course of its adaptation to compensate for loss of DNA ligase (40). Alterations in gene 3 were previously observed in T7 phage that suppresses the loss of the ligase gene, gene 1.3, when grown on a ligase-deficient host (41). These studies suggested that T7 phage modifies genes whose products are involved in DNA replication and/or recombination in order to adapt to alterations in an essential gene. However, since the enzymatic activity of some proteins is agonistic or antagonistic to other proteins, compensatory modification of genes must be precisely regulated in order to achieve effective overall function. For example, we found that mutations in gene 3 that suppress the phenotype of gp5-4N arise when gene 3 is expressed from phage. When both gene 3 and gene 5 are overexpressed from supplied plasmids, compensation of phage growth by mutated gene 3 is not observed (data not shown). This observation supports the suggestion that compensatory changes need to be balanced in a DNA metabolism network (3, 41). We have also observed mutations in gene 3 that can suppress defects in T7 phage that have mutations in other replication proteins such as gene 2.5 ssDNA binding protein (B. Marintcheva, unpublished results) or gene 4 helicase-primase (S.-J. Lee, unpublished results).
Although alteration of the basic region in T7 gp5 clearly results in DNA polymerase of low processivity, it is still elusive if the low processivity is solely responsible for reduction in its ability to complement T7 phage lacking gene 5. Considering the complexity of the replisome, the altered gp5 could have other defects that involve interactions with other replication proteins. Indeed, we have observed that gp5-4N/Trx does not properly interact with gene 4 helicase to carry out strand displacement synthesis (S.-J. Lee, unpublished results).
Acknowledgments
This work was supported by Public Health Service Grant GM 54397 from National Institutes of Health.
We thank Steve Moskowitz for preparing the figures.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59219-228. [DOI] [PubMed] [Google Scholar]
- 2.Brieba, L. G., B. F. Eichman, R. J. Kokoska, S. Doublie, T. A. Kunkel, and T. Ellenberger. 2004. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 233452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull, J. J., and I. J. Molineux. 2008. Predicting evolution from genomics: experimental evolution of bacteriophage T7. Heredity 100453-463. [DOI] [PubMed] [Google Scholar]
- 4.Center, M. S., and C. C. Richardson. 1970. An endonuclease induced after infection of Escherichia coli with bacteriophage T7. I. Purification and properties of the enzyme. J. Biol. Chem. 2456285-6291. [PubMed] [Google Scholar]
- 5.Center, M. S., F. W. Studier, and C. C. Richardson. 1970. The structural gene for a T7 endonuclease essential for phage DNA synthesis. Proc. Natl. Acad. Sci. USA 65242-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 40437-41. [DOI] [PubMed] [Google Scholar]
- 8.Declais, A. C., J. Hadden, S. E. Phillips, and D. M. Lilley. 2001. The active site of the junction-resolving enzyme T7 endonuclease I. J. Mol. Biol. 3071145-1158. [DOI] [PubMed] [Google Scholar]
- 9.Doublie, S., M. R. Sawaya, and T. Ellenberger. 1999. An open and closed case for all polymerases. Structure 7R31-R35. [DOI] [PubMed] [Google Scholar]
- 10.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391251-258. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166477-535. [DOI] [PubMed] [Google Scholar]
- 12.Dutta, S., Y. Li, D. Johnson, L. Dzantiev, C. C. Richardson, L. J. Romano, and T. Ellenberger. 2004. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc. Natl. Acad. Sci. USA 10116186-16191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endy, D., L. You, J. Yin, and I. J. Molineux. 2000. Computation, prediction, and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc. Natl. Acad. Sci. USA 975375-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engler, M. J., and C. C. Richardson. 1983. Bacteriophage T7 DNA replication. Synthesis of lagging strands in a reconstituted system using purified proteins. J. Biol. Chem. 25811197-11205. [PubMed] [Google Scholar]
- 15.Guan, C., and S. Kumar. 2005. A single catalytic domain of the junction-resolving enzyme T7 endonuclease I is a non-specific nicking endonuclease. Nucleic Acids Res. 336225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadden, J. M., M. A. Convery, A. C. Declais, D. M. Lilley, and S. E. Phillips. 2001. Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I. Nat. Struct. Biol. 862-67. [DOI] [PubMed] [Google Scholar]
- 17.Hadden, J. M., A. C. Declais, S. B. Carr, D. M. Lilley, and S. E. Phillips. 2007. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature 449621-624. [DOI] [PubMed] [Google Scholar]
- 18.Hamdan, S. M., B. Marintcheva, T. Cook, S. J. Lee, S. Tabor, and C. C. Richardson. 2005. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. USA 1025096-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himawan, J. S., and C. C. Richardson. 1992. Genetic analysis of the interaction between bacteriophage T7 DNA polymerase and Escherichia coli thioredoxin. Proc. Natl. Acad. Sci. USA 899774-9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollis, T., J. M. Stattel, D. S. Walther, C. C. Richardson, and T. Ellenberger. 2001. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. USA 989557-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmgren, A., B. O. Soderberg, H. Eklund, and C. I. Branden. 1975. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 Å resolution. Proc. Natl. Acad. Sci. USA 722305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong, G., and K. N. Kreuzer. 2003. Endonuclease cleavage of blocked replication forks: an indirect pathway of DNA damage from antitumor drug-topoisomerase complexes. Proc. Natl. Acad. Sci. USA 1005046-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hori, K., D. F. Mark, and C. C. Richardson. 1979. Deoxyribonucleic acid polymerase of bacteriophage T7. Characterization of the exonuclease activities of the gene 5 protein and the reconstituted polymerase. J. Biol. Chem. 25411598-11604. [PubMed] [Google Scholar]
- 24.Huber, H. E., M. Russel, P. Model, and C. C. Richardson. 1986. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. The redox capacity of thioredoxin is not required for stimulation of DNA polymerase activity. J. Biol. Chem. 26115006-15012. [PubMed] [Google Scholar]
- 25.Huber, H. E., S. Tabor, and C. C. Richardson. 1987. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 26216224-16232. [PubMed] [Google Scholar]
- 26.Johnson, D. E., and C. C. Richardson. 2003. A covalent linkage between the gene 5 DNA polymerase of bacteriophage T7 and Escherichia coli thioredoxin, the processivity factor: fate of thioredoxin during DNA synthesis. J. Biol. Chem. 27823762-23772. [DOI] [PubMed] [Google Scholar]
- 27.Kim, Y. T., and C. C. Richardson. 1993. Bacteriophage T7 gene 2.5 protein: an essential protein for DNA replication. Proc. Natl. Acad. Sci. USA 9010173-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman, New York, NY.
- 29.Lee, J., P. D. Chastain II, T. Kusakabe, J. D. Griffith, and C. C. Richardson. 1998. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell 11001-1010. [DOI] [PubMed] [Google Scholar]
- 30.Lilley, D. M., and M. F. White. 2001. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2433-443. [DOI] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.McGlynn, P., R. G. Lloyd, and K. J. Marians. 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 988235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Modrich, P., and C. C. Richardson. 1975. Bacteriophage T7 deoxyribonucleic acid replication in vitro. A protein of Escherichia coli required for bacteriophage T7 DNA polymerase activity. J. Biol. Chem. 2505508-5514. [PubMed] [Google Scholar]
- 35.Molineux, I. 2006. The T7 group, p. 277-301. In R. Calendar (ed.), The bacteriophages, 2nd ed., Oxford University Press, New York, NY.
- 36.Myers, T. W., and L. J. Romano. 1988. Mechanism of stimulation of T7 DNA polymerase by Escherichia coli single-stranded DNA binding protein (SSB). J. Biol. Chem. 26317006-17015. [PubMed] [Google Scholar]
- 37.Parkinson, M. J., J. R. Pohler, and D. M. Lilley. 1999. Catalytic and binding mutants of the junction-resolving enzyme endonuclease I of bacteriophage T7: role of acidic residues. Nucleic Acids Res. 27682-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pomerantz, R. T., and M. O'Donnell. 2007. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 15156-164. [DOI] [PubMed] [Google Scholar]
- 39.Richardson, C. C. 1983. Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell 33315-317. [DOI] [PubMed] [Google Scholar]
- 40.Rokyta, D., M. R. Badgett, I. J. Molineux, and J. J. Bull. 2002. Experimental genomic evolution: extensive compensation for loss of DNA ligase activity in a virus. Mol. Biol. Evol. 19230-238. [DOI] [PubMed] [Google Scholar]
- 41.Sadowski, P. D. 1974. Suppression of a mutation in gene 3 of bacteriophage T7 (T7 endonuclease I) by mutations in phage and host polynucleotide ligase. J. Virol. 13226-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Melocular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Singleton, M. R., M. R. Sawaya, T. Ellenberger, and D. B. Wigley. 2000. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101589-600. [DOI] [PubMed] [Google Scholar]
- 44.Tabor, S., H. E. Huber, and C. C. Richardson. 1987. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 26216212-16223. [PubMed] [Google Scholar]
- 45.Thrall, B. D., D. B. Mann, M. J. Smerdon, and D. L. Springer. 1992. DNA polymerase, RNA polymerase and exonuclease activities on a DNA sequence modified by benzo[a]pyrene diolepoxide. Carcinogenesis 131529-1534. [DOI] [PubMed] [Google Scholar]
- 46.Yang, X. M., and C. C. Richardson. 1997. Amino acid changes in a unique sequence of bacteriophage T7 DNA polymerase alter the processivity of nucleotide polymerization. J. Biol. Chem. 2726599-6606. [DOI] [PubMed] [Google Scholar]