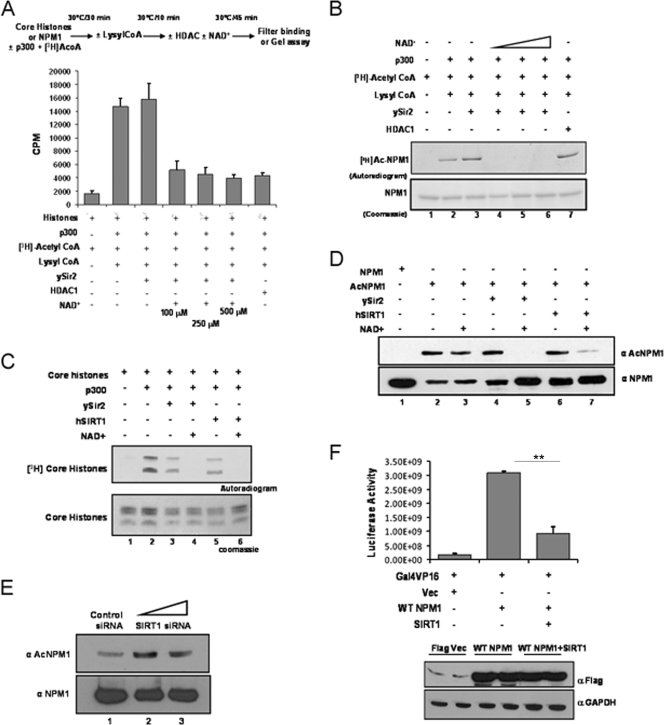
FIG. 3.
hSIRT1 deacetylates NPM1. (A) Results of a filter binding assay done to normalize the HDAC activities of ySir2 and HDAC1 with core histones (purified from HeLa cells) as the substrate are represented as bar graphs. The details of the reaction are given as a schematic (upper panel). The p300 inhibitor lysyl coenzyme A (CoA) was added at the end of the acetylation step to prevent reacetylation of proteins in the subsequent deacetylation reaction. (B) Acetylated (by p300) NPM1 was subjected to deacetylation with ySir2 with or without NAD+ and HDAC1. Unlabeled NPM1 (lane 1), 3H-labeled acetylated NPM1 (lane 2), acetylated NPM1 incubated with ySir2 (lane 3), acetylated NPM1 incubated with ySir2 in the presence of increasing concentrations of NAD+ (lanes 4 to 6), and acetylated NPM1 incubated with HDAC1 (lane 7) are shown. (C) Gel picture depicting the normalization of HDAC activity of ySir2 and hSIRT1 with core histones (purified from HeLa cells) as the substrate. (D) Acetylated NPM1 was incubated with the indicated deacetylases with or without NAD+, followed by Western blotting with anti-AcNPM1-01. A corresponding loading control is shown after reprobing of the same blot with anti-NPM1 antibody. (E) A Western blot analysis of SIRT1 siRNA with anti-AcNPM1 antibody was done at 48 h posttransfection (upper panel). A Western blot analysis with anti-NPM1 antibody was done to show the corresponding loading control (lower panel). (F) A luciferase assay was done at 48 h posttransfection with 1 μg of either the Flag vector (Vec) alone (lane 1) or Flag-tagged WT NPM1 (lane 2) and also upon the cotransfection of SIRT1 with Flag-tagged WT NPM1 (lane 3) in HEK293T cells along with pG10luc (100 ng), Gal4-VP16 (10 ng), and pCMV-gal (100 ng). Values are means ± standard deviations of triplicates. The data were analyzed by paired t test (**, P < 0.01). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.