Abstract
The AP-2 transcription factor family is linked with development of the head and limbs in both vertebrate and invertebrate species. Recent evidence has also implicated this gene family in the evolution of the neural crest in chordates, a critical step that allowed the development and elaboration of the vertebrate craniofacial skeleton. In mice, the inappropriate embryonic expression of one particular AP-2 gene, Tcfap2a - encoding AP-2α - results in multiple developmental abnormalities, including craniofacial and limb defects. Thus, Tcfap2a provides a valuable genetic resource to analyze the regulatory hierarchy responsible for the evolution and development of the face and limbs. Previous studies have identified a 2 kilobase intronic region of both the mouse and human AP-2α locus that directs expression of a linked LacZ transgene to the facial processes and the distal mesenchyme of the limb bud in transgenic mice. Further analysis identified two highly conserved regions of ~ 200-400 bp within this tissue-specific enhancer. We have now initiated a transgenic and biochemical analysis of the most important of these highly conserved regions. Our analysis indicates that although the sequences regulating face and limb expression have been integrated into a single enhancer, different cis-acting sequences ultimately control these two expression domains. Moreover, these studies demonstrate that a conserved STAT binding site provides a major contribution to the expression of Tcfap2a in the facial prominences.
Keywords: Tcfap2a, limb bud, frontal nasal process, STAT1, Sox6
Introduction
The mature mammalian face is formed by the fusion of seven facial primordia (Helms et al., 2005): the frontal nasal process (FNP), paired lateral nasal processes (LNPs), paired maxillary processes (MxPs), and paired mandibular processes (MnPs). The cellular and molecular events underlying craniofacial formation are complex, but there is a growing appreciation that the growth and patterning of the facial primordia has many parallels with development of the limb buds (Mariani and Martin, 2003; Niswander, 2003; Tickle, 2003; Helms et al., 2005). The genetic defects responsible for several human craniofacial syndromes are often accompanied by limb defects (Wilkie and Morriss-Kay, 2001; Ornitz and Marie, 2002; Thyagarajan et al., 2003; Zelzer and Olsen, 2003; Chen and Deng, 2005; L'Hote and Knowles, 2005), and mutations in a number of mouse genes can also cause morphological alterations in both the face and the limbs (Richman and Lee, 2003; Helms et al., 2005). In molecular terms, the two developmental systems rely on a combination of sonic hedgehog (shh) and fibroblast growth factor (FGF) signals to impart both survival and polarity information to the face and limbs. (Hu et al., 2003; Mariani and Martin, 2003; Niswander, 2003; Richman and Lee, 2003; Tickle, 2003; Abzhanov and Tabin, 2004; Helms et al., 2005). Similarly, many transcription factors are utilized during both face and limb development, and mouse models with disrupted expression of these transcription factors often result in both craniofacial and limb defects. Such proteins include AP-2α, Twist, Cbfa1/Runx2, and members of the retinoic acid receptor, Gli, Sox, Msx, Dlx, and Alx transcription factor families (Richman and Lee, 2003).
AP-2α is a member of a small family of “basic-helix-span-helix” transcription factors important for many aspects of vertebrate development (Schorle et al., 1996; Zhang et al., 1996; Moser et al., 1997; Nottoli et al., 1998; Hilger-Eversheim et al., 2000; Satoda et al., 2000; Auman et al., 2002; Werling and Schorle, 2002; Mani et al., 2005). The transcription factor AP-2α, encoded by Tcfap2a in mouse and TFAP2A in human, is expressed in both the developing face and limbs (Mitchell et al., 1991). Mice bearing a homozygous deletion of Tcfap2a die perinatally and have severe defects in many distinct morphogenetic processes including limb and craniofacial development (Schorle et al., 1996; Zhang et al., 1996). Craniofacial defects include hypoplastic growth or complete loss of cranial neural crest cell (CNCC) derivatives and midline and lateral clefting. The majority of Tcfap2a null mice exhibit zeugopod defects in the forelimb (Schorle et al., 1996; Zhang et al., 1996). Studies using Tcfap2a-/- ↔ Tcfap2a+/+ chimeric mice, as well as the analysis of mice containing tissue specific deletions of Tcfap2a, have reinforced the importance of this transcription factor for face and limb development (Nottoli et al., 1998; Brewer et al., 2004; Nelson and Williams, 2004). In these latter experiments, the altered patterns of AP-2α expression resulted in a spectrum of craniofacial defects, including cleft lip with or without cleft palate, isolated cleft secondary palate, and reduced suture growth, while limb defects included syndactyly and polysyndactyly (Nottoli et al., 1998; Brewer et al., 2004; Nelson and Williams, 2004).
Given the strong association between AP-2α and the growth and development of the face and limbs we have begun to identify the cis-acting sequences and trans-acting factors required for the expression of this transcription factor in these tissues during their development. We have previously identified a 2 Kb enhancer encompassing the fifth intron of TFAP2A responsible for directing AP-2α expression to the developing FNP, LNPs, and limb bud mesenchyme (LBM) in transgenic mice (Zhang and Williams, 2003). In this report, we sought to link expression of AP-2α to upstream signaling pathways by assessing the organization of the FNP/LBM-specific enhancer. Since the AP-2 transcription factor family has been implicated in the evolution of neural crest in chordates (Meulemans and Bronner-Fraser, 2002; Luo et al., 2003; Meulemans and Bronner-Fraser, 2004), a critical step that has allowed the development and elaboration of the vertebrate craniofacial skeleton, studies on Tcfap2a should provide valuable insight into the regulatory hierarchy responsible for the evolution of the face and limbs. Herein, we utilized a transgenic approach to identify minimal sub-fragments of the enhancer that are required for expression in both the FNP and the LBM. In addition, we performed biochemical analyses of the minimal enhancer elements and identify both Sox factor and STAT binding sites that contribute to the expression of Tcfap2a in the FNP. Identification of these sites provides insight into the pathways that act upstream of AP-2α in the FNP and will provide the foundation for further dissection of the role that AP-2α plays in craniofacial growth, development, and evolution.
Results
Expression of Tcfap2a in the Developing FNP and LBM Is Mediated by a Highly Conserved Element
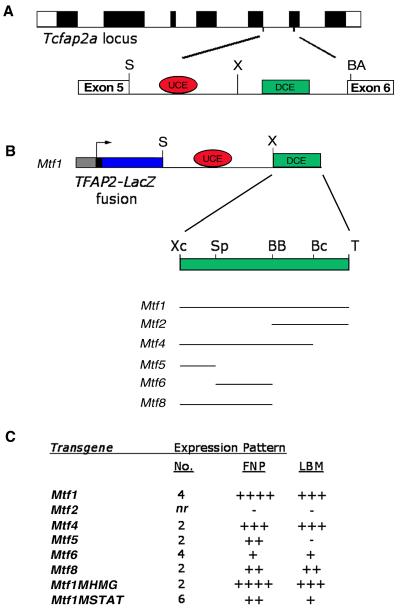
Our previous studies have localized an FNP/LBM-specific enhancer to intron 5 of both the human and mouse gene locus encoding AP-2α (Zhang and Williams, 2003; Zhang et al., manuscript in preparation). Within this 2 kb region, there are two distinct conserved regions, a moderately conserved upstream region of 200 bp (Fig. 1, red oval, UCE; upstream conserved element) and a more highly conserved downstream region of 400 bp (Fig. 1, green rectangle, DCE; downstream conserved element). However, the relative importance of these conserved sequences, as well as other regions within the enhancer, remain untested. Therefore, we have sought to identify both the minimal enhancer necessary for directing expression to the face and limbs as well as the individual elements within the enhancer that mediate control over Tcfap2a expression. For this purpose we utilized the minimal human AP-2α promoter attached to LacZ (Zhang and Williams, 2003) to develop a series of transgenes into which specific portions of the mouse intronic enhancer were then inserted (Fig. 1). In a prior experiment we had found that when we divided the intron into two sections only the downstream 1 kb region of the enhancer containing the DCE could direct tissue-specific expression to the FNP and LBM. The more upstream 1kb fragment containing the UCE did not direct LacZ expression alone in vivo, although it enhanced expression driven by the region containing the DCE (Zhang et al., manuscript in preparation). Therefore, for the current analysis, we generated a modified construct in which just the 400 bp DCE (Fig. 1B, XC-T: XcmI-TaqI) was combined with the 1kb fragment containing the UCE and subsequently linked to the TFAP2A-LacZ transgene (Fig. 1B). This minimized transgenic construct, Mtf1 (Fig. 1B and C), recapitulated the expression profile of Tcfap2a in the face at E10.5, including the FNP, the LNPs, and a medial region at the caudal margin of the maxillary prominences (Figs 2A, B; also see Fig. 5A). Similarly, appropriate expression was observed in the LBM, but as expected this element did not direct expression to the AER in transgenic mice (Figs 2A, C). These data indicate that Mtf1, which comprises the 1kb upstream fragment containing the UCE, in combination with just the downstream DCE region of the Tcfap2a fifth intron, could direct robust tissue-specific expression to the face and limbs. Therefore we utilized this minimal transgene as the basis for all subsequent constructs.
Fig. 1. Transgenic analysis of the Tcfap2a FNP/LBM enhancer.
(A) Schematic representation of the Tcfap2a locus with exons depicted as filled boxes. Intron 5 is expanded to highlight two subregions of high sequence conservation. The larger, distal conserved element (DCE, green rectangle) is essential for expression in the FNP and LBM. The upstream conserved element (UCE, red oval) enhances the expression of the DCE. (B) Schematic representation of the transgenic constructs utilized in these studies. Each construct consisted of 200 bps of TFAP2A basal promoter elements, which direct the expression of the Tcfap2a-LacZ fusion transgene (including 13 amino acids of AP-2α). The enhancer elements include the SpeI-XhoI UCE (red oval) from intron 5 and variable fragments of the DCE (green rectangle). The DCE is expanded to show subfragments defined by restriction enzyme sites. Abbreviations: S, SpeI; X, XhoI; BA, BsaAI; Xc, XcmI; Sp, SphI; BB, BstBI; Bc, BclI; and T, TaqI. (C) Summary of expression data acquired from each transgene. For each construct the number (No.) of transgenic embryos sharing the same pattern of expression is indicated. Expression was scored in the FNP and the LBM. Relative intensity and domain of expression is indicated by (+), while (-) indicates that no expression was observed in the transgenic embryos. The notation “nr” indicates that the number of transgenic embryos that failed to give specific staining was not recorded, but was greater than 2.
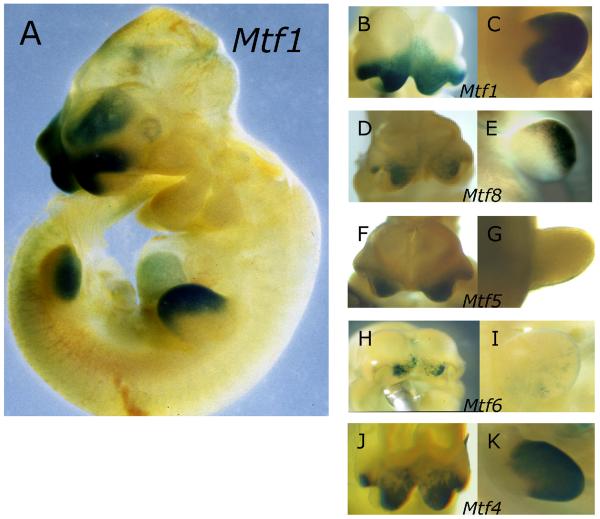
Fig. 2. Expression of β-galactosidase in transgenic embryos.
(A) Lateral view of an E10.5 Mtf1 transgenic embryo showing specific expression in the FNP and LBM. Detailed ventral views of FNP (B, D, F, H, J) and lateral views of forelimb buds C, E, G, I, K) from embryos transgenic for Mtf1 (B, C), Mtf8 (D, E), Mtf5 (F, G) Mtf6 (H, I), and Mtf4 (J, K).
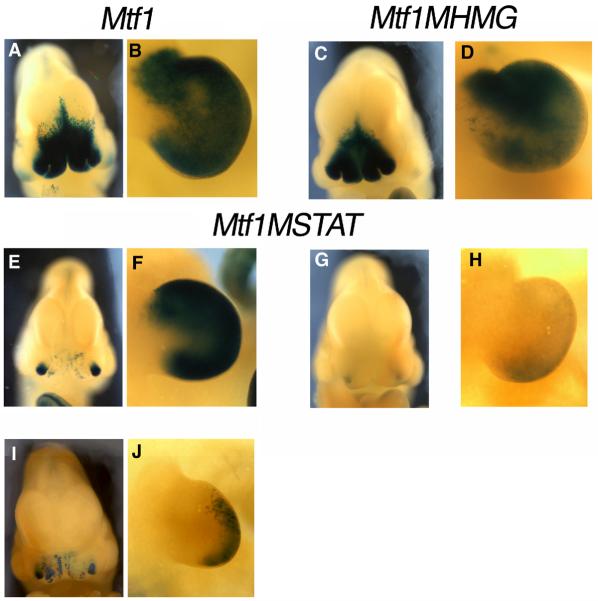
Fig. 5. The STAT site is important for the in vivo activity of the DCE in the FNP.
Images of ß-galactosidase stained E10.5 embryos transgenic for Mtf1 (A, B), Mtf1MHMG (C, D) or Mtf1MSTAT (E-J). Frontal view of the facial prominences of embryos transgenic for Mtf1 (A), Mtf1MHMG (C) or Mtf1MSTAT (E, G, I). Lateral view of the forelimb bud of Mtf1 (B), Mtf1MHMG (D), and three Mtf1MSTAT (F, H, J) embryos.
The DCE Controls Expression in the FNP and LBM Differently
To map subregions of the DCE that are responsible for expression in the FNP and LBM, we divided the DCE into two sub-fragments and these were then inserted into the TFAP2A-LacZ transgenic construct alongside the 1kb UCE fragment to generate either Mtf2 or Mtf8 (Fig. 1B). Mtf8, containing the more upstream half of the DCE, directed expression of the transgene to the FNP and the LBM (Figs 2D, E). The expression observed in the facial prominences from this construct, however, was consistently weaker than the expression derived from Mtf1 (compare Figs 2B, D). Likewise, the expression in the LBM was weaker for Mtf8 than for Mtf1 (compare Figs 2C, E). On the other hand, Mtf2, possessing the more 3' portion of the DCE, did not direct any specific expression of the transgene (data not shown).
Together these transgenic experiments show that elements necessary for AP-2α expression in the FNP and LBM are contained within the first 200 bps of the DCE. These data also indicated that elements located in the 3' half of the DCE (Mtf2) might augment tissue-specific activity, since the level of expression derived from Mtf8 alone is less than that of the entire element, Mtf1. To test this hypothesis, we generated the construct Mtf4 (Fig. 1B), which contained the Mtf8 region and an additional 100 bps from Mtf2 (BB-Bc: BstBI to BclI). Like Mtf8, Mtf4 directed expression to the FNP and LBM (Figs 2J, K), but again the expression in the FNP was weaker than observed from Mtf1 (Figs 2B, J). In contrast, expression in the LBM of Mtf4 embryos was greater than that observed for Mtf8 and similar to the level of observed for Mtf1 (compare Figs 2C, E, K). These findings show that the majority of cis-regulatory sequences necessary for the appropriate level and distribution of limb-specific expression are contained within the upstream 300 bp of the DCE, whereas face-specific expression is derived from elements located throughout the DCE.
To define the minimal region necessary for Tcfap2a expression, we subdivided the DCE portion present in Mtf8 into two smaller ~ 100bp fragments. Again these subfragments were attached to the TFAP2A-LacZ transgene along with the UCE to generate Mtf5 and Mtf6 (Fig. 1B), and these constructs were tested in transgenic mice. Mtf5, like Mtf8, was capable of directing expression of the transgene to the FNP (Fig. 2F). Unlike Mtf8, however, Mtf5 did not direct any expression to the LBM (Fig. 2G). Mtf6 also directed weak expression to the FNP and only minor patchy expression to the LBM (Figs 2H, I). The intensity of expression in the FNP observed in Mtf5 embryos was weaker than that observed for Mtf8 embryos (Figs 2D, F), while expression in Mtf6 embryos was significantly weaker (Figs 2D, H).
Thus, minimal elements necessary for Tcfap2a expression in the FNP are located in the 5' end of the DCE. It should be noted, however, that expression was obtained in the FNP with two completely separate 100 bp fragments from the DCE, corresponding to the Mtf5 and Mtf6 transgenes, indicating that at least two distinct regions are capable of directing minimal expression of Tcfap2a to the FNP. In the limbs, on the other hand, only the entire Mtf8 portion of the DCE yielded robust expression, indicating that elements contained within, or overlapping between, Mtf5 and Mtf6 must be combined to direct expression to the limbs. This finding is significant, since it demonstrates that the regulation of Tcfap2a in the FNP and the LBM is not identical.
In total, these data indicate that there are elements within each subdomain of the DCE that contribute to the expression of Tcfap2a in the FNP. The bulk of the expression is derived from the 5' region of the DCE, contained within the Mtf5 transgene. Addition of incremental 100 bp fragments of the DCE to generate Mtf8, Mtf4, and Mtf1 respectively, each yields greater transgene expression in the FNP. Regulation in the limbs, however, does not follow this same pattern. Mtf8 was the smallest transgene to yield significant expression in the LBM. This expression is enhanced by the addition of a 100 bp of the DCE to yield Mtf4. The final 3' DCE sub-region within Mtf1 does not alter the intensity of expression in the limbs.
Sox Factors Bind the DCE In Vitro
We next utilized a biochemical approach to identify potentially important cis-acting elements within the DCE. Given the high degree of sequence conservation between the mouse, human, and chick enhancers in this region, as well as the similarity of endogenous AP-2α expression in the mouse and chick face (Mitchell et al., 1991; Shen et al., 1997), the analysis was initiated using whole cell protein extracts derived from the chick FNP mesenchyme. Differences in the mechanisms of embryogenesis between mouse and chick, particularly the contrast between in utero versus in ovo development, make the chick a more efficient and cost-effective system for an initial analysis of the DNA binding proteins present in the early face. Subsequently, we were able to employ the data obtained using chick extracts for a focused analysis of the transcription factors present in the more limited mouse head extracts (see below).
To begin these experiments, we first generated the construct pCK2-4, which contains the entire DCE sequence, but is derived from the equivalent chicken region rather than the mouse region contained within the Mtf1 transgenic construct. Next, we utilized the chick sequences for DNase I footprinting experiments in the presence of whole cell protein extracts derived from Stage 25 chick FNP mesenchyme (Fig. 3A). At this stage of chick embryogenesis, FNP development approximates that observed at E10.5 in mouse, the stage at which transgene expression was assessed. In the presence of increasing concentrations of chick FNP extract, several subtle changes were observed in the pattern of DNase I cleavage, with the most dramatic change being the creation of a hypersensitive site in the upstream part of the DCE (Fig. 3A, arrow). A band just above the hypersensitive site was also potentially protected from cleavage in the presence of extracts (Fig. 3A, arrowhead).
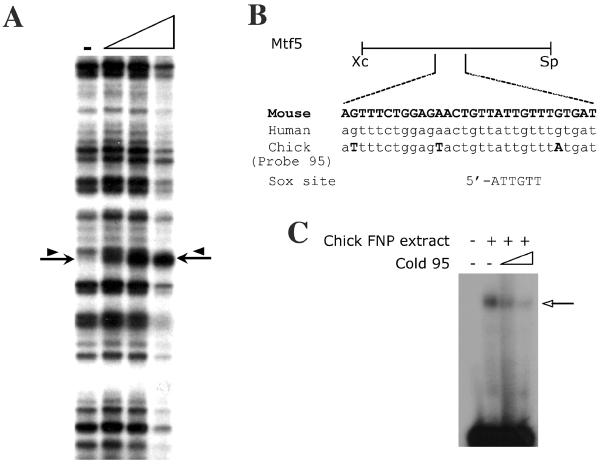
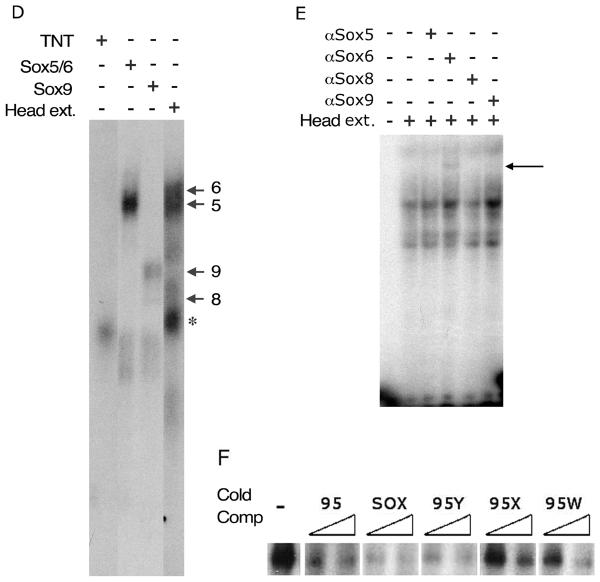
Fig. 3. Sox factors interact with the Tcfap2a face and limb enhancer.
(A). DNase I footprint on CK2-4 with protein extracts from Stage 25 chick FNP extracts. A DNase I digested lane that lacks extract (-) is shown at the left and the remaining tracks have increasing amounts of FNP extract. The position of the DNase I hypersensitive site (arrow) and the band that potentially becomes protected by extract (arrowhead) are indicated. The region affected in the CK2-4 footprint corresponds to Probe 95. (B) Schematic representation of the sub-region of the DCE FNP/LBM-specific enhancer present in the Mtf5 transgene. The sequence of Probe 95 (chick sequence) is shown along with the alignment of the corresponding regions from human and mouse. A Sox consensus sequence is aligned with the putative Sox binding site in Probe 95. (C) EMSAs with radiolabeled Probe 95 in the presence of protein extracts from Stage 25 chick FNP (arrow indicates DNA-protein complex formation). Competition with increasing quantities of cold Probe 95 indicates that the binding is specific. (D). EMSAs in the presence of mouse head extracts or in vitro translated Sox5, Sox6, or Sox9 on Probe 95. The arrows indicate the relative positions of the DNA complexes formed by each individual Sox protein (this panel and data not shown). The Sox5/6 shift comigrates with a complex from the mouse head extracts. A non-specific shift from the reticulocyte extracts is indicated by *. (E). EMSAs with anti-Sox5, -Sox6, -Sox8, or -Sox9 antibodies in the presence of protein extracts from E10.5 mouse heads. The super-shift in the presence of the anti-Sox6 antibody is indicated (arrow). (F) Competition analysis for Sox6 bound to Probe 95 in the absence (-) or presence of increasing concentrations of cold competitors as indicated (see Table 1A for sequences).
The sequence surrounding the DNase I hypersensitive site is shown in Fig. 3B. To confirm that a factor in the chick FNP extracts interacts with this sequence, we performed Electophoretic Mobility Shift Assays (EMSAs) with a radiolabeled probe, Probe 95, derived from this region (Fig. 3B). In the presence of extracts from chick FNP mesenchyme, a single protein-DNA complex was observed (Fig. 3C, arrow). This complex was competed successfully by the introduction of excess unlabeled Probe 95 (Fig. 3C). These data indicate that the region identified in the footprint analysis, corresponding to Probe 95, binds specifically to a factor in the chick FNP extracts. Analysis of the sequence in this region revealed an SRY-related HMG box (Sox) binding site (5'-AACAAT-3') (Mertin et al., 1999). Several members of the Sox gene family are required for face and limb development, particularly for skeletogenesis, and so represented important potential candidates for Tcfap2a regulation in these structures (Nakashima and de Crombrugghe, 2003; Richman and Lee, 2003; Sock et al., 2004). To determine whether Sox factors are indeed capable of interacting with this element we generated full-length L-Sox5, Sox6, Sox8, and Sox9 proteins by in vitro transcription and translation. These factors were chosen since they are representative of the Sox genes expressed in both the developing FNP and LBM (Lefebvre et al., 1998; Wegner, 1999). We observed robust and specific protein:DNA complexes in the presence of Sox5, Sox6, or Sox9 programmed extracts, and a weaker interaction in the presence of Sox8 (Fig. 3D and data not shown). Sox6 generated a slightly slower migrating band than Sox5, with Sox9 and Sox8 generating sequentially faster migrating complexes, consistent with the sizes of the proteins produced by in vitro transcription and translation. These complexes were distinct from any non-specific bands produced by the un-programmed rabbit reticulocyte lysate (Fig 3D, asterisk). Furthermore, all specific Sox protein:probe DNA complexes were abolished when the non-specific competitor poly (dG-dC) was replaced with poly (dI-dC), a compound that is known to compete for Sox protein DNA binding (data not shown). These findings indicate that members of the Sox protein family which are expressed in the mouse face and limb can interact with the DNA sequences encompassing the hypersensitive site within the FNP/LBM enhancer of the Mtf5 subfragment of the DCE.
We next utilized whole cell protein extracts from E10.5 mouse heads in EMSAs to identify specific protein factors present in the mouse embryo that interact with this region of the enhancer. In the presence of mouse head extract, several protein-DNA complexes were observed, and one of these migrated in close proximity to bands generated by Sox5/6 generated in vitro (Fig. 3D). The mouse extract was then pre-incubated with antisera specific for Sox5, Sox6, Sox8, and Sox9 protein prior to EMSA (Fig. 3E). The presence of anti-Sox6 antibody generated a new super-shifted complex (Fig. 3E, lane 4, arrow), whereas none of the other antisera tested produced a super-shifted complex (Fig. 3E). These experiments indicate that Sox6 may be uniquely present at sufficient concentrations in E10.5 head extracts for detection in these assays. An alternative possibility is that only the Sox6 antiserum had the ability to recognize the endogenous mouse protein within the extract as well as produce a supershifted complex.
Next, we wanted to identify sequences within the FNP enhancer that are critical for Sox6 interaction. For these experiments, we generated mutant probes, 95W, 95X, 95Y, and 95Z (Table 1A) and compared their ability to bind Sox6 with either probe 95 or a standard Sox binding site. 95X contained a G to C mutation at position 4 of the Sox binding site within probe 95 - a substitution that has previously been shown to interfere with Sox factor-DNA interaction (Mertin et al., 1999). 95W also contains this substitution as well as a second change in the consensus Sox site (Table 1A). Neither Probe 95X or 95W were as efficient as Probe 95 or the Sox binding site when used as cold competitor oligonucleotides in EMSA reactions containing Sox6 and radiolabeled Probe 95 (Fig. 3F). These findings support the ability of the 5'-ATTGTT-3' motif we have identified to act as a bona fide Sox binding site. We also noted that probe 95 contained an adjacent sequence that closely resembled a second possible Sox binding site (5'-ACTGTT-3') (Table 1A). We therefore tested the ability of the 95Y sequence, which has a mutation in just this adjacent Sox-like site, to act as a competitor. Probe 95Y was approximately as efficient as a cold competitor sequence as Probe 95 or the consensus Sox site (Fig. 3F). We next tested probe 95Z, containing both the 95X mutation in the bona fide Sox consensus sequence as well as the 95Y alteration in the adjacent sequence, and determined that this behaved similarly to the 95X sequence alone in Sox6 binding assays (data not shown). These results indicate that the adjacent Sox-like sequence does not contribute significantly to the ability of Probe 95 to bind Sox6 either alone or via cooperative interactions with the cognate Sox consensus sequence. In summary, using both DNAseI footprinting and EMSA we have identified a conserved region of the Tcfap2a FNP/LBM enhancer, 5'-ATTGTT-3', that can interact with proteins present in both mouse and chick embryonic head extracts. This sequence resides within the Mtf5 fragment of the DCE and can bind Sox proteins, particularly Sox6, both in vitro and in vivo.
Table 1.
Sequences of Probes, Consensus Sequences, and Transgenic Mutations
| A. Oligo name | Sequence |
|---|---|
| 95 | 5′-ATTTTCTGGAGTACTGTTATTGTTTATGAT |
| SOX | 5′-ATTGTT |
| 95W | 5′-ACTGTTCTTCTT |
| 95X | 5′-ACTGTTATTCTT |
| 95Y | 5′-ACTCTTATTGTT |
| 95Z | 5′-ACTCTTATTCTT |
| MHMG | 5′-ACTGTACTTCTT |
| B. Oligo name | Sequence |
|---|---|
| 45 | 5′-CAACTGGTTCAGGAAATTAAAACC |
| STAT | 5′-TTCCGGAA |
| ETS | 5′-AGCCGGAA |
| bHLH | 5′-CACGTG |
| MSTAT | 5′-AACCAACTGGTGCACGCTATTAAAACCAAAGATT |
Stat1 Interacts with the DCE In Vitro
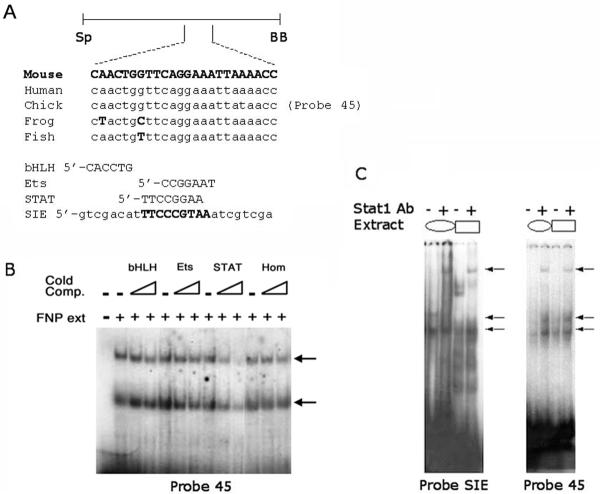
Despite the absence of an obvious DNase I footprint in the sequences encompassing the Mtf6 portion of the DCE we initiated a more thorough EMSA analysis of this region of the enhancer because it augments expression in both the face and limbs and is also weakly capable of directing expression to the FNP by itself. The 100 bp region was subdivided into several overlapping oligonucleotides and these were used for EMSAs in combination with extracts from the chick FNP mesenchyme (Fig. 4 and data not shown). One of these oligonucleotides, Probe 45, formed two major specific protein-DNA complexes in the presence of the chicken FNP extracts (Figs 4A and B). Sequence analyses of Probe 45 identified several putative protein binding sites including: a basic-helix-loop-helix site; an Ets binding site; and a STAT recognition element (Fig. 4A and Table 1B) (Quandt et al., 1995). Since the region of the DCE present in Mtf6 was highly A/T rich, homeodomain proteins might also interact with these sequences. To determine which of these classes of transcription factor were interacting with Probe 45, we performed competitive EMSAs using radiolabeled Probe 45 and extracts from the chick FNP. We introduced cold, competitor oligonucleotides containing consensus sequences for each of the aforementioned classes of transcription factors (see Table 1B). The cold competitor containing a STAT recognition element was uniquely able to compete for the complexes formed in the presence of chick extract (Fig. 4B). Interestingly, the competitor containing the Ets binding site, which had very similar sequence (Table 1B), was unable to affect the formation of these complexes (Fig. 4B). These data suggest that one or more Stat proteins (Signal Transducers and Activators of Transcription) bind this sequence in vitro.
Fig. 4. Stat1 binds to the Mtf6 DCE region of the Tcfap2a FNP/LBM enhancer.
(A) Schematic representation of the Mtf6 DCE subfragment of the FNP/LBM enhancer. The sequence of Probe 45, a highly conserved sub-fragment of the Mtf6 DCE region, contains several putative binding sites for known classes of transcriptional regulators including; bHLH, Ets, STAT, and homeodomain proteins. Consensus sequences for these putative binding factors are shown. SIE is a naturally occurring binding site for Stat1. (B) EMSA experiment using radiolabelled Probe 45 either with (+) or without (-) protein extract from Stage 25 chick FNP. The two specific complexes formed are indicated (arrows). Lanes also contained either no specific cold competitor DNA (-) or various cold competitors containing particular consensus sequences (listed in A and indicated above the relevant lanes) at 10-fold and 100-fold excess in adjacent lanes. Only the cold competitor containing the STAT consensus competed effectively for the upper shift. (C) Stat1 in extracts from U-937 cells treated with IFNa (oval) and E10.5 mouse head extracts (rectangle) binds to both SIE and Probe 45 (lower arrows). The Stat1 shifts on both probes and with both types of extracts are super-shifted in the presence of a Stat1-specific antibody (upper arrows).
In order to unequivocally demonstrate that Stat factors can interact with Probe 45, we performed EMSAs with extracts from either IFNa-treated human U937 monocytic cells, a source of phosphorylated Stat proteins, or E10.5 mouse heads (Fig. 4C). EMSAs were performed on radiolabeled Probe 45 and a control, Probe SIE (Stat inducible element). Protein-DNA complexes were observed on Probe SIE with extracts from both U937 cells and E10.5 mouse head. Although, as expected, the complexes formed by these two extracts were not identical, both generated a new slowly migrating, super-shifted complex in the presence of a Stat1-specific antiserum (Fig. 4C). These experiments indicate that Stat1 is indeed present in our E10.5 mouse head extracts. The same experiments were performed on Probe 45. In the presence of extracts from both U937 cells and E10.5 mouse heads, two protein-DNA complexes were observed, and an additional super-shifted complex was obtained following pre-incubation with the Stat1-specific antiserum (Fig. 4C). Thus, Stat1 is present in extracts from E10.5 mouse heads and is capable of interacting with Probe 45 in vitro.
Stat Proteins Regulate Tcfap2a In Vivo
Next, we wanted to determine whether the conserved Sox and STAT binding sites we had identified were important for the in vivo activity of the FNP/LBM enhancer. For these experiments, we generated two modified Mtf1 transgenic constructs. Mtf1MHMG was identical to Mtf1, except that it contained the 95W mutations within the Sox6 binding site that greatly impacted Sox6 binding (Fig. 3F; Table 1A). Surprisingly, Mtf1MHMG transgenic embryos had normal expression of the LacZ transgene in both the FNP and LBM when compared with the non-mutated Mtf1 transgene (Fig. 5). These results demonstrate that mutation of this single conserved Sox binding site alone is insufficient to disrupt the in vivo activity of the FNP/LBM-specific enhancer.
The second modified transgenic construct we generated, Mtf1MSTAT, was identical to Mtf1, except that the core sequence of the STAT binding site in the Mtf6 DCE fragment, 5'-TTCAGGAA-3', was altered to 5'-TGCACGCT-3' (Table 1B). Like Mtf1MHMG, the Mtf1MSTAT transgene still directed b-galactosidase expression to the face and limb buds. However, for this latter transgene the levels of expression were greatly reduced compared to that observed with the parental Mtf1 transgene (Fig. 5). Out of six Mtf1MSTAT transgenic embryos that displayed ß-galactosidase activity, five gave weak to moderate activity in the FNP, while only three had weak to moderate activity in the LBM (Fig. 5 and data not shown). These data indicate that the loss of the Stat binding site greatly impacts the function of the FNP/LBM enhancer. Residual activity was still observed in the FNP, however, and we hypothesized that this was because the intact Mtf5 fragment, which alone can drive FNP expression, was present within the Mtf1MSTAT transgene (Fig. 2). To address this issue we generated a modified Mtf6 transgene, Mtf6MSTAT, which had a mutated STAT binding site and lacked the Mtf5 fragment of the DCE. Multiple injections with this Mtf6MSTAT construct failed to produce transgenic embryos with any detectable b-galactosidase activity in the FNP or the LBM (data not shown). Thus, the STAT binding site within the Mtf6 portion of the DCE is the major cis-acting sequence within this region required for in vivo enhancer activity in the FNP and LBM.
Discussion
Regulation of Tcfap2a in the FNP and LBM Are Controlled by Different Mechanisms From the Same Enhancer
The AP-2α transcription factor is critical for many aspects of mouse craniofacial development, including neural crest induction, neural tube closure, fusion of the facial prominences, and the formation of the secondary palate (Schorle et al., 1996; Zhang et al., 1996; Nottoli et al., 1998; Brewer et al., 2004; Nelson and Williams, 2004). The human gene encoding AP-2α, TFAP2A, maps in close proximity to a region of chromosome 6p24 that is frequently involved in translocations and deletions associated with orofacial clefting (Davies et al., 1999a; Davies et al., 1999b; Topping et al., 2002; Davies et al., 2004; Schultz et al., 2004) - raising the possibility that alterations in the long range cis-regulatory sequences controlling TFAP2A expression might be responsible for congenital defects affecting human facial development. In the context of evolution, evidence from multiple species indicates that changes in the spatial and temporal regulation of the AP-2 family of genes, among others, might underlie the evolution of the vertebrate neural crest and ultimately the diversity of craniofacial skeleton (Meulemans and Bronner-Fraser, 2002; Holzschuh et al., 2003; Knight et al., 2003; Luo et al., 2003; O'Brien et al., 2004; Knight et al., 2005). To identify the regulatory hierarchy underlying the expression of Tcfap2a, and to determine how it might have been altered during evolution, or in human genetic disease, we have previously identified regions of the gene encoding AP-2α from various species that are responsible for driving the expression of Tcfap2a in different tissues (Zhang and Williams, 2003; Zhang et al., manuscript in preparation). Here, we have focused on one of these elements, located within the fifth intron of Tcfap2a, that directs expression to the developing face and limbs. We were particularly interested in this element because gene expression and function in the limbs and face are often linked. Indeed, it has been postulated that development of the face shares many regulatory circuits in common with appendages such as the limbs and genitalia (Yamaguchi et al., 1999; Yamada et al., 2003; Klonisch et al., 2004; Cobb and Duboule, 2005). In this report, we have shown that two conserved regions of the intron work together to yield high levels of expression in the face and limbs. The more extensive downstream element (DCE) is responsible for the tissue-specific activity in the face and limbs, and within this sequence we have identified minimal elements required for expression in each tissue. While elements responsible for expression in both tissues overlap, the minimal requirements for the two tissues are not identical. Minimal expression in the FNP can be obtained from two distinct 100 bp fragments located at the 5' end of the DCE, present within the transgenes Mtf5 and Mtf6. The combination of these two subfragments in Mtf8 yields more robust expression in the FNP, while the complete pattern of expression in the FNP is only achieved when the entire 400 bp DCE region is present in the Mtf1 transgene.
Minimal significant expression in the LBM, on the other hand, is obtained from only the intact upstream 200 bp region of the DCE present in Mtf8, and the addition of a further downstream region of 100 bp (Mtf4) is needed to restore robust LBM activity. These findings suggest that elements that are each capable of directing expression to the FNP must be combined to direct expression to the LBM. In addition, these studies show that elements within the 3' half of the enhancer contribute differently to the intensity of expression in the FNP and the LBM. Thus, while elements that control expression in the FNP and LBM overlap, they are not identical.
We note that our data may also support a model in which particular regions of the DCE differentially regulate subtle aspects of the spatial pattern of FNP or LBM expression as well as exerting an influence on the overall level of expression. This possibility is most strongly supported by a comparison of the Mtf1 and Mtf1MSTAT transgenic data. The intact Mtf1 transgene directs similar expression to both the lateral and medial nasal prominences. In contrast, Mtf1MSTAT yields only patchy expression in the medial facial prominences, whereas expression in the lateral prominences is more robust and concentrated (Fig. 5). Therefore, as well as affecting the overall levels of Tcfap2a expression throughout the FNP, the Mtf6 DCE region and its associated Stat binding site may exert a more profound effect on expression within the medial nasal prominences. The integration of cis-regulatory elements controlling different domains of gene expression into this Tcfap2a enhancer may serve as a model for how subtle changes in the organization and arrangement of DNA binding sites within an enhancer can affect morphogenesis and patterning during evolution and development.
Sox Proteins Interact with the Tcfap2a Enhancer Element
We have provided compelling evidence that Sox6 interacts with a Sox binding site within the DCE of Tcfap2a. The Sox site is located in a subdomain of the DCE that is necessary for strong Tcfap2a expression in the FNP and LBM (Fig. 1C and Fig. 6). We have shown that chick FNP extracts interacting with the Sox binding site in DNase I footprinting assays create a prominent hypersensitive site. Such hypersensitive sites often result from protein-DNA interactions that dramatically alter the structure of DNA, as when Sox factors introduce significant bends into DNA (van Houte et al., 1995; Werner et al., 1995; Murphy et al., 2001). Additionally, several Sox family members, including Sox6, can bind to the enhancer in EMSAs, and we have shown that Sox6 is present in mouse head extracts and can interact with the enhancer. The alteration of this Sox site alone, though, in the transgene Mtf1MHMG, is not sufficient to produce a significant change in the activity of the Mtf1 enhancer element in vivo. There are several possible explanations for this result. First, although the mutation that we introduced significantly reduced the affinity of Sox6 for site in vitro, the residual binding potential may be sufficient to direct expression in vivo. This is especially likely if cooperative complex formation between Sox6 and a co-factor could augment interaction with a weak site. Cooperative binding between Sox proteins and co-factors is typically required for the activity and specificity of Sox proteins (reviewed in (Kamachi et al., 2000; Wilson and Koopman, 2002)). Indeed, one possible class of partners are the Sox proteins themselves as several weaker Sox binding sites are predicted to occur within the DCE based on the presence of the core binding sequence 5'-TTGTT-3' (Fig. 6, Table 1A). An alternative possibility is that the Sox binding site has a minimal role in the context of the isolated enhancer for the FNP/LBM during the developmental window we have analyzed, but is important in the wider context of endogenous Tcfap2a expression. Further analyses will be required to determine the importance of this Sox binding site for the other tissues, such as the skin and mammary gland, in which this enhancer cooperates with more distal sequences to direct expression (Zhang and Williams, 2003; Zhang et al., manuscript in preparation).
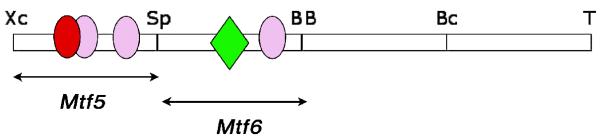
Fig. 6. Summary of the Sox and STAT binding sites in the DCE.
A representative summary of binding sites located in the DCE. The position of bona fide Sox site within probe 95 that was revealed by DNase I footprint analysis (red oval), several putative Sox factor binding sites (pink ovals), and the STAT binding site in probe 45 (green diamond) are shown.
Although we only have direct evidence that Sox6 is present in the facial extracts, it is likely that more than one Sox protein will be able to interact with the Sox DCE element in vivo. Several Sox factors are known to play critical roles in craniofacial and limb skeletogenesis and are good candidates to regulate Tcfap2a (Bi et al., 1999; Bi et al., 2001; Smits et al., 2001; Sock et al., 2004). The Group E factor, Sox9, is an enticing candidate, given both its role in chondrogenesis and its responsiveness to retinoic acid, (Bi et al., 1999; Sekiya et al., 2000; Bi et al., 2001; Akiyama et al., 2002; Mori-Akiyama et al., 2003; Akiyama et al., 2004; Eames et al., 2004; Sahar et al., 2005) an agent known to alter AP-2α expression in vitro and in vivo (Williams et al., 1988; Lüscher et al., 1989; Philipp et al., 1994; Shen et al., 1997). Another Group E candidate, Sox8, is expressed coincident with Tcfap2a in the face and limbs (Wegner, 1999; Schepers et al., 2000). Mouse Sox8 can also regulate osteoblast differentiation, while SOX8 maps to a chromosomal locus implicated in the human syndrome ATR-16, which has associated craniofacial anomalies (Pfeifer et al., 2000; Schmidt et al., 2005).
Two Group C family members, L-Sox5 and Sox6, are expressed in the face and limbs coincident with Tcfap2a (Lefebvre et al., 1998). Homozygous mutation of Sox5 in mice produces a perinatal lethal phenotype involving respiratory distress, cleft palate, and delayed mineralization of the nasal bones (Smits et al., 2001). Sox6 null mice also die shortly after birth from respiratory distress (Smits et al., 2001). A few Sox6 null embryos survive for two weeks and they rapidly develop severe dwarfism (Smits et al., 2001). Sox5/Sox6 double knockout embryos have round heads and severely shortened snouts by E13.5 and exhibit severe chondrodysplasia (Smits et al., 2001). Sox6, in particular, is an enticing upstream regulator of Tcfap2a since premature differentiation of osteoblasts was observed at the nasal bone front in FKO mice, which lack AP-2α in the FNP (Nelson and Williams, 2004), and Sox6 is the only one of the three Sox chondrogenic markers (Sox5, Sox6, and Sox9) to also be expressed during intramembranous bone formation (Eames and Helms, 2004). Recently, the Group C Sox gene, Sox11, has also been shown to influence craniofacial development. Mice lacking Sox11 frequently showed unilateral and bilateral orofacial clefts that were similar to phenotypes observed with certain Tcfap2a mutations ((Sock et al., 2004) and W. Feng and T. Williams, unpublished). Further studies, however, are needed to prove which Sox family member(s) act upstream of Tcfap2a.
STAT Proteins Act Upstream of Tcfap2a
We have identified a highly conserved STAT binding site in the region of the Tcfap2a FNP/LBM DCE enhancer contained within the Mtf6 transgene (Fig. 6). Mutation of the STAT site in the transgene Mtf1MSTAT greatly reduces FNP and LBM-specific enhancer activity associated with the DCE, and eliminates it completely in the context of the Mtf6 transgene alone. Thus, the STAT site is a critical component of the Mtf6 DCE region required for appropriate FNP/LBM activity of the fifth intron enhancer sequence. Further, we have shown that the Stat1 protein is present in our mouse head extracts and can bind to this element in vitro. While Stat1 is one possible candidate for regulating Tcfap2a expression, we note that several other Stat family members with similar DNA binding specificities are also expressed in the developing facial mesenchyme or skeletal elements, including Stat3, 5a, and 5b (Duncan et al., 1997). Thus, although the STAT binding site itself is critical for expression, there may be functional redundancy amongst the Stat proteins for binding and regulation of this Tcfap2a cis-acting sequence. In this context, preliminary studies indicate that expression driven by Tcfap2a is not significantly altered in Stat1 null mice (S. Sullivan and T. Williams, unpublished observations). Nevertheless, as a whole, the members of the Stat family of proteins are excellent candidates to act as upstream regulators of Tcfap2a given their developmental roles in skeletogenesis as well as their biochemical function as downstream effectors of FGF signal transduction.
Homozygous deletion of Stat3, in mice, results in early embryonic lethality (Takeda et al., 1997), and thus, the contribution of Stat3 to craniofacial and limb bud development has not been assessed. Bones formed via either endochondral or intramembranous ossification in Stat1-/- mice have an increase in mass and mineral density due to a disruption in the proliferation and differentiation properties of both osteoblasts and osteoclasts (Kim et al., 2003; Xiao et al., 2004), and FGFs have an antiproliferative effect on chondrocytes that is disrupted by loss of Stat1 (Sahni et al., 1999; Sahni et al., 2001).
Stat-1, -3, -5a, and -5b are each activated downstream of FGFR3 during endochondral bone growth. FGFR3 activation leads to phosphorylation and nuclear localization of Stat-1, -5a, and -5b in chondrocytes (Su et al., 1997; Li et al., 1999), while Stat3 is activated by FGFR3 in cell culture (Hart et al., 2000). FGFR3 mutations in humans cause a spectrum of achondroplasias (reviewed in (Ornitz and Marie, 2002; Chen and Deng, 2005; L'Hote and Knowles, 2005)). Mouse models for the FGFR3-related achondroplasias demonstrate that FGFR3 negatively regulates post-natal long bone growth, while constitutive activation of FGFR3 causes dwarfism (Ornitz and Marie, 2002; Chen and Deng, 2005; L'Hote and Knowles, 2005). FGFR3 related skeletal defects arise from both altered proliferation of chondrocytes in the growth plate and premature ossification (Ornitz and Marie, 2002; Chen and Deng, 2005; L'Hote and Knowles, 2005; Ornitz, 2005). In contrast to the general inhibitory effect of FGFR3 on post-natal long bone growth, a SADDAN (severe achondroplasia with developmental delay and acanthosis nigricans) FGFR3 mouse model exhibited overgrowth of the nasal septum and a high incidence of dental malocclusion (Iwata et al., 2001). These phenotypes are reminiscent of defects observed in FKO and Tcfap2a+/- mice (Zhang et al., 1996; Nottoli et al., 1998; Nelson and Williams, 2004).
Stat proteins are also activated downstream of other FGFRs (Hart et al., 2000). Mutations in both FGFR1 and -2 have been implicated in craniofacial syndromes, but are more commonly found in craniosynostosis, premature fusion of sutures due to disrupted intramembranous bone growth (reviewed in (Chen and Deng, 2005)). Although an in vivo role for Stats downstream of FGFR2 has not yet been shown, mouse models of craniosynostosis containing altered FGFR2, display several phenotypes that overlap with those observed in FKO, Wnt1-Cre; Floxdel/KI, or Tcfap2a+/- mice including shortened snouts, dental malocclusion, wide spaced eyes, and cleft palate (Zhang et al., 1996; Nottoli et al., 1998; De Moerlooze et al., 2000; Hajihosseini et al., 2001; Eswarakumar et al., 2002; Chen et al., 2003; Brewer et al., 2004; Eswarakumar et al., 2004; Nelson and Williams, 2004; Wang et al., 2005). Thus, the presence of a STAT site in the FNP/LBM-specific enhancer likely places AP-2α downstream of FGFR signaling in the FNP, which is consistent with other studies showing FGF dependent changes of Tcfap2a in the craniofacial region (Shen et al., 1997; Cordero et al., 2004).
In summary, we have identified minimal enhancer elements that control Tcfap2a expression in both the FNP and LBM. Identification of these minimal elements aided in the discovery of binding sites that control Tcfap2a expression in the FNP and its derivatives and may contribute to Tcfap2a expression in the LBM. While the specific Sox and Stat factors that interact with the Tcfap2a FNP/LBM-specific enhancer remain to be determined, these results link Tcfap2a to known signaling pathways and further implicate AP-2α in the regulation of bone growth and skeletal development.
Experimental Procedures
Plasmids
CK2-4 was amplified by PCR from chicken genomic DNA using the primers CK2 (5'-CTC CGT TTA GCT ACG GAG CA-3') and CK4 (5'-CAT AGC CAC GTG CAT ACG-3'). PCR reactions contained 180 ng of chicken genomic DNA, 1 mg of each CK2 and CK4, and Taq DNA polymerase at conditions recommended by the manufacturer (Boehringer Mannheim). The resulting 545 bp fragment was subcloned into pBSKS+ (Stratagene). Subfragments of the DCE for transgenic constructs were generated by PCR, as above, from mouse genomic DNA and were subcloned into pBSKS (Stratagene) to generate shuttle plasmids (i.e. Mts1). Primers used to create DNA fragments were: Mts1, 5'-XcmI (5'-ATG TGG TGG AAG CTG AGG GTA G-3') and 3'-TaqI (5'- CCC GAC TGC AG TTT CCT CG-3'); Mts2, 5'BstBI (5'-CGA AAA TCA ACA ACA CAG ACA GAC-3') and 3'-TaqI; Mts4, 5'XcmI and 3'BclI (5'-GCG AAC AAC CGA GAA AGT TTA C-3'); Mts5, 5'-XcmI and 3'-SphI (5'-CGC CAA TTA GAG CAT CAA AGC-3'); Mts6, 5'-SphI (5'-GAT GCT CTA ATT GGC GCA TGC-3') and 3'-BstBI (5'-CTG TCT GTG TTG TTG ATT TTC G-3'); Mts8, 5'-XcmI and 3'-BclI. All shuttle plasmids were converted to final transgenic (i.e. Mtf1) constructs by three-way ligation of the Mts XhoI/NotI subfragment, a 1 Kb SpeI/XhoI fragment containing the UCE, and a large SpeI/NotI fragment containing the TFAP2A-LacZ fusion transgene. Mtf1MHMG, Mtf1MSTAT, and Mtf6MSTAT were generated by PCR mutagenesis of Mtf1 or Mtf6 (for sequences, see Table 1) according to manufacturer instructions (Stratagene). All plasmids were sequenced at the Keck Foundation sequencing facility (Yale University).
Protein Extracts
Fertilized chick eggs (Charles River-SPAFAS) were harvested at Stage 25 (Hamburger and Hamilton, 1951). FNP mesenchyme and ectoderm were microdissected with tungsten needles. FNP tissue was soaked in 2% trypsin (Sigma) for 20 minutes at 25°C. Ectoderm was teased away from mesenchymal buds and discarded. Buds of mesenchyme tissue were pooled in ice cold 1X PBS; 10% Fetal Calf Serum. Mesenchyme was pipetted repeatedly to dissociate tissue. Chick FNP extracts were prepared as described (Andrews and Faller, 1991). Mouse extracts were generated from 200 E10.5 mouse heads as described (Shashikant et al., 1995). Extracts were dialysed against 3 changes of Modified Buffer A (50 mM Tris pH 7.6; 5 mM MgCl2; 25 mM KCl; 0.2 mM EDTA; 10% glycerol).
Sox proteins were generated with TNT® Quick Coupled Transcription/Translation systems according to manufacturer instructions (Promega).
Electrophoretic Mobility Shift Assays
Oligonucleotides for EMSA probes were synthesized at the Keck Foundation at Yale University. Complimentary oligonucleotides were annealed in 10 mM Tris, pH 7.9; 5 mM MgCl2; 50 mM NaCl by heating at 95°C for 5 minutes and cooling slowly to 4°C. Probes for EMSA were 5'-end labeled using γ[32P] ATP (New England Nuclear) with T4 Polynucleotide Kinase (New England Biolabs). Labeled probes were separated from unincorporated ATP using Centri-spin 20 columns (Princeton Separations). EMSA reactions contained 10,000 cpm probe, 1X EMSA Buffer (10% glycerol; 5 mM MgCl2; 2.5 mM EDTA; 2.5 mM DTT; 250 mM NaCl; 50 mM Tris, pH 7.5), and either 1 μg poly (dI-dC) or poly (dG-dC). Reactions were incubated for 30 minutes on ice prior to resolution via 6% native PAGE. EMSAs to detect Stat1-specific binding with IFNα-treated U-937 cells were performed with a NuShift™ Stat1a kit according to manufacturer instructions (Active Motif). Antisera to other Stat proteins were obtained from Zymed Laboratories, Santa Cruz Biotechnology, and Geneka Biotechnology, but only Stat1 antiserum generated a specific supershift (data not shown). The Sox6 antiserum was a gift of Véronique Lefebvre. Super-shift EMSAs were completed by preincubating antibodies with extracts for 1 hour at 4°C prior to addition of radiolabeled probe. DNase I footprints were performed as described (Williams et al., 1988).
Generation and Staining of Transgenic Mouse Embryos
All animal experiments were performed in accordance with protocols approved by the UCHSC or Yale University Animal Care and Usage Committees. Transgenic FVB mice were generated by standard methods (Nagy et al., 2003). Embryos were collected from foster females (CD-1) at E10.5, with the day of the transgenic manipulation being scored E0.5. The actual developmental stage of the embryos varied due to ex vivo manipulation (Nagy et al., 2003). Embryos were stained for β-galactosidase activity by standard procedures (Nagy et al., 2003), fixed in 4% paraformaldehyde, and photographed. Staining was standardized to 18 hours to facilitate a direct comparison of expression amongst the different transgenes.
Acknowledgements
We are grateful to Véronique Lefebvre, Benoit de Crombrugghe, Michael Wegner and Peter Koopman for providing assistance with Sox reagents. ALD is grateful to Stephanie Donaldson for training in transgenic mouse generation. The authors thank members of the Williams laboratory for helpful discussions and assistance, and Kristin Artinger for critical reading gof the manuscript. ALD was supported by a Sessel Anonymous fellowship (Yale University) and an NRSA from NIDCR (F32-DE05735). The research was supported by the NIDCR grant R01 DE12728 (T.W.).
Grant Sponsor: NIDCR; Grant number: DE12728
References
- Abzhanov A, Tabin CJ. Shh and Fgf8 act synergistically to drive cartilage outgrowth during cranial development. Dev Biol. 2004;273:134–148. doi: 10.1016/j.ydbio.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and β-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. Transcription factor AP-2γ is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129:2733–2747. doi: 10.1242/dev.129.11.2733. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre mediated deletion of AP-2α causes multiple neural crest related defects. Dev Biol. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Chen L, Deng CX. Roles of FGF signaling in skeletal development and human genetic diseases. Front Biosci. 2005;10:1961–1976. doi: 10.2741/1671. [DOI] [PubMed] [Google Scholar]
- Chen L, Li D, Li C, Engel A, Deng CX. A Ser250Trp substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone. 2003;33:169–178. doi: 10.1016/s8756-3282(03)00222-9. [DOI] [PubMed] [Google Scholar]
- Cobb J, Duboule D. Comparative analysis of genes downstream of the Hoxd cluster in developing digits and external genitalia. Development. 2005;132:3055–3067. doi: 10.1242/dev.01885. [DOI] [PubMed] [Google Scholar]
- Cordero D, Marcucio R, Hu D, Gaffield W, Tapadia M, Helms JA. Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J Clin Invest. 2004;114:485–494. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AF, Mirza G, Flinter F, Ragoussis J. An interstitial deletion of 6p24-p25 proximal to the FKHL7 locus and including AP-2α that affects anterior eye chamber development. Journal of Medical Genetics. 1999a;36:708–710. [PMC free article] [PubMed] [Google Scholar]
- Davies AF, Mirza G, Sekhon G, Turnpenny P, Leroy F, Speleman F, Law C, van Regemorter N, Vamos E, Flinter F, Ragoussis J. Delineation of two distinct 6p deletion syndromes. Human Genetics. 1999b;104:64–72. doi: 10.1007/s004390050911. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Wise C, Venkatesh B, Mirza G, Jefferson A, Volpi EV, Ragoussis J. Mapping of three translocation breakpoints associated with orofacial clefting within 6p24 and identification of new transcripts within the region. Cytogenet Genome Res. 2004;105:47–53. doi: 10.1159/000078008. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Zhong Z, Wen Z, Darnell JE., Jr. STAT signaling is active during early mammalian development. Dev Dyn. 1997;208:190–198. doi: 10.1002/(SICI)1097-0177(199702)208:2<190::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Eames BF, Helms JA. Conserved molecular program regulating cranial and appendicular skeletogenesis. Dev Dyn. 2004;231:4–13. doi: 10.1002/dvdy.20134. [DOI] [PubMed] [Google Scholar]
- Eames BF, Sharpe PT, Helms JA. Hierarchy revealed in the specification of three skeletal fates by Sox9 and Runx2. Dev Biol. 2004;274:188–200. doi: 10.1016/j.ydbio.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Horowitz MC, Locklin R, Morriss-Kay GM, Lonai P. A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc Natl Acad Sci U S A. 2004;101:12555–12560. doi: 10.1073/pnas.0405031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129:3783–3793. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- Hajihosseini MK, Wilson S, De Moerlooze L, Dickson C. A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc Natl Acad Sci U S A. 2001;98:3855–3860. doi: 10.1073/pnas.071586898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of Morphology. 1951;88:49–92. 1951. [PubMed] [Google Scholar]
- Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000;19:3309–3320. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- Helms JA, Cordero D, Tapadia MD. New insights into craniofacial morphogenesis. Development. 2005;132:851–861. doi: 10.1242/dev.01705. [DOI] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. 2000;260:1–12. doi: 10.1016/s0378-1119(00)00454-6. [DOI] [PubMed] [Google Scholar]
- Holzschuh J, Barrallo-Gimeno A, Ettl AK, Durr K, Knapik EW, Driever W. Noradrenergic neurons in the zebrafish hindbrain are induced by retinoic acid and require tfap2a for expression of the neurotransmitter phenotype. Development. 2003;130:5741–5754. doi: 10.1242/dev.00816. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- Iwata T, Li CL, Deng CX, Francomano CA. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Hum Mol Genet. 2001;10:1255–1264. doi: 10.1093/hmg/10.12.1255. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonisch T, Fowler PA, Hombach-Klonisch S. Molecular and genetic regulation of testis descent and external genitalia development. Dev Biol. 2004;270:1–18. doi: 10.1016/j.ydbio.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development. 2005;132:3127–3138. doi: 10.1242/dev.01879. [DOI] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch G-J, Schilling TF. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- L'Hote CG, Knowles MA. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Exp Cell Res. 2005;304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. Embo J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2α. Proc Natl Acad Sci U S A. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B, Mitchell PJ, Williams T, Tjian R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and by second messengers. Genes Dev. 1989;3:1507–1517. doi: 10.1101/gad.3.10.1507. [DOI] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Farhi A, Carew KS, Warnes CA, Nelson-Williams C, Day RW, Pober B, State MW, Lifton RP. Syndromic patent ductus arteriosus: evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc Natl Acad Sci U S A. 2005;102:2975–2979. doi: 10.1073/pnas.0409852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Martin GR. Deciphering skeletal patterning: clues from the limb. Nature. 2003;423:319–325. doi: 10.1038/nature01655. [DOI] [PubMed] [Google Scholar]
- Mertin S, McDowall SG, Harley VR. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Timmons PM, Hebert JM, Rigby PWJ, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci U S A. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Pscherer A, Roth C, Becker J, Mucher G, Zerres K, Dixkens C, Weis J, Guay-Woodford L, Buettner R, Fassler R. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2β. Genes and Development. 1997;11:1938–1948. doi: 10.1101/gad.11.15.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EC, Zhurkin VB, Louis JM, Cornilescu G, Clore GM. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J Mol Biol. 2001;312:481–499. doi: 10.1006/jmbi.2001.4977. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2003. [Google Scholar]
- Nakashima K, de Crombrugghe B. Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet. 2003;19:458–466. doi: 10.1016/S0168-9525(03)00176-8. [DOI] [PubMed] [Google Scholar]
- Nelson DK, Williams T. Frontonasal process-specific disruption of AP-2α results in postnatal midfacial hypoplasia, vascular anomalies, and nasal cavity defects. Dev Biol. 2004;267:72–92. doi: 10.1016/j.ydbio.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Niswander L. Pattern formation: old models out on a limb. Nat Rev Genet. 2003;4:133–143. doi: 10.1038/nrg1001. [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, Williams T. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proc Natl Acad Sci U S A. 1998;95:13714–13719. doi: 10.1073/pnas.95.23.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien EK, d'Alencon C, Bonde G, Li W, Schoenebeck J, Allende ML, Gelb BD, Yelon D, Eisen JS, Cornell RA. Transcription factor AP-2α is necessary for development of embryonic melanophores, autonomic neurons and pharyngeal skeleton in zebrafish. Dev Biol. 2004;265:246–261. doi: 10.1016/j.ydbio.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Ornitz DM. FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 2005;16:205–213. doi: 10.1016/j.cytogfr.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Poulat F, Holinski-Feder E, Kooy F, Scherer G. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics. 2000;63:108–116. doi: 10.1006/geno.1999.6060. [DOI] [PubMed] [Google Scholar]
- Philipp J, Mitchell PJ, Malipiero U, Fontana A. Cell type-specific regulation of expression of transcription factor AP-2 in neuroectodermal cells. Dev Biol. 1994;165:602–614. doi: 10.1006/dbio.1994.1279. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman JM, Lee SH. About face: signals and genes controlling jaw patterning and identity in vertebrates. Bioessays. 2003;25:554–568. doi: 10.1002/bies.10288. [DOI] [PubMed] [Google Scholar]
- Sahar DE, Longaker MT, Quarto N. Sox9 neural crest determinant gene controls patterning and closure of the posterior frontal cranial suture. Dev Biol. 2005;280:344–361. doi: 10.1016/j.ydbio.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13:1361–1366. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni M, Raz R, Coffin JD, Levy D, Basilico C. STAT1 mediates the increased apoptosis and reduced chondrocyte proliferation in mice overexpressing FGF2. Development. 2001;128:2119–2129. doi: 10.1242/dev.128.11.2119. [DOI] [PubMed] [Google Scholar]
- Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, Davidson HR, Pierpont ME, Gelb BD. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet. 2000;25:42–46. doi: 10.1038/75578. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Bullejos M, Hosking BM, Koopman P. Cloning and characterisation of the Sry-related transcription factor gene Sox8. Nucleic Acids Res. 2000;28:1473–1480. doi: 10.1093/nar/28.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Schinke T, Haberland M, Priemel M, Schilling AF, Mueldner C, Rueger JM, Sock E, Wegner M, Amling M. The high mobility group transcription factor Sox8 is a negative regulator of osteoblast differentiation. J Cell Biol. 2005;168:899–910. doi: 10.1083/jcb.200408013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Schultz RE, Cooper ME, Daack-Hirsch S, Shi M, Nepomucena B, Graf KA, O'Brien EK, O'Brien SE, Marazita ML, Murray JC. Targeted scan of fifteen regions for nonsyndromic cleft lip and palate in Filipino families. Am J Med Genet. 2004;125A:17–22. doi: 10.1002/ajmg.a.20424. [DOI] [PubMed] [Google Scholar]
- Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is upregulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- Shashikant CS, Bieberich CJ, Belting HG, Wang JC, Borbely MA, Ruddle FH. Regulation of Hoxc-8 during mouse embryonic development: identification and characterization of critical elements involved in early neural tube expression. Development. 1995;121:4339–4347. doi: 10.1242/dev.121.12.4339. [DOI] [PubMed] [Google Scholar]
- Shen H, Wilkie T, Ashique AM, Narvey M, Zerucha T, Savino E, Williams T, Richman JM. Chicken transcription factor AP-2: cloning, expression and its role in outgrowth of facial promineneces and limb buds. Dev Biol. 1997;188:248–266. doi: 10.1006/dbio.1997.8617. [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Croumbrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WC, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton WA, Fu XY. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan T, Totey S, Danton MJ, Kulkarni AB. Genetically altered mouse models: the good, the bad, and the ugly. Crit Rev Oral Biol Med. 2003;14:154–174. doi: 10.1177/154411130301400302. [DOI] [PubMed] [Google Scholar]
- Tickle C. Patterning systems--from one end of the limb to the other. Dev Cell. 2003;4:449–458. doi: 10.1016/s1534-5807(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Topping A, Harris P, Moss AL. The 6p deletion syndrome: a new orofacial clefting syndrome and its implications for antenatal screening. British Journal of Plastic Surgery. 2002;55:68–72. doi: 10.1054/bjps.2001.3729. [DOI] [PubMed] [Google Scholar]
- van Houte LP, Chuprina VP, van der Wetering M, Boelens R, Kaptein R, Clevers H. Solution structure of the sequence-specific HMG box of the lymphocyte transcriptional activator Sox-4. J Biol Chem. 1995;270:30516–30524. doi: 10.1074/jbc.270.51.30516. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Yang F, Karim BO, Iacovelli AJ, Cai J, Lerner CP, Richtsmeier JT, Leszl JM, Hill CA, Yu K, Ornitz DM, Elisseeff J, Huso DL, Jabs EW. Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development. 2005;132:3537–3548. doi: 10.1242/dev.01914. [DOI] [PubMed] [Google Scholar]
- Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling U, Schorle H. Transcription factor gene AP-2g essential for early murine development. Mol Cell Biol. 2002;22:3149–3156. doi: 10.1128/MCB.22.9.3149-3156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Williams T, Admon A, Lüscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Xiao L, Naganawa T, Obugunde E, Gronowicz G, Ornitz DM, Coffin JD, Hurley MM. Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J Biol Chem. 2004;279:27743–27752. doi: 10.1074/jbc.M314323200. [DOI] [PubMed] [Google Scholar]
- Yamada G, Satoh Y, Baskin LS, Cunha GR. Cellular and molecular mechanisms of development of the external genitalia. Differentiation. 2003;71:445–460. doi: 10.1046/j.1432-0436.2003.7108001.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Olsen BR. The genetic basis for skeletal diseases. Nature. 2003;423:343–348. doi: 10.1038/nature01659. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Williams T. Identification and Regulation of Tissue-Specific cis-acting Elements Associated with the Human AP-2α Gene. Dev Dyn. 2003;228:194–207. doi: 10.1002/dvdy.10365. [DOI] [PubMed] [Google Scholar]