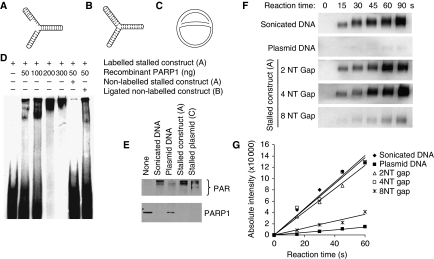
Figure 2.
PARP1 binds to and is activated by DNA fork structures in vitro. (A) Biotin-labelled stalled fork construct and (B) ligated construct, containing sealed DNA ends. (C) Early replication intermediate of the plasmid pBROTB535, containing replication forks stalled in vitro by omission of topoisomerase from the replication reaction. (D) Electrophoretic mobility shift assay using biotin-labelled artificial stalled fork substrate and increasing concentrations of purified PARP1 protein with or without a 10-fold excess of non-labelled competitor stalled fork substrate or ligated construct. (E) Western blot analysis of PARP1 (bottom) and PAR (top) after incubation of 50 ng purified PARP protein with 50 ng of different DNA substrates. Automodification reduces the electrophoretic mobility of PARP1, accounting for the decreased amounts of unmodified PARP1 protein detectable at its expected molecular size in these samples. (F) PARP1 activation by increasing length of gap within the stalled fork structure (A). Recombinant human PARP1 (5 nM) was incubated with DNA constructs and biotinylated NAD+ for the times indicated, and blots were probed with anti-biotin antibody. Sonicated DNA was used as positive control and plasmid DNA as negative control. (G) Quantification of PARP1 activation as in (F).