Fig. 8.
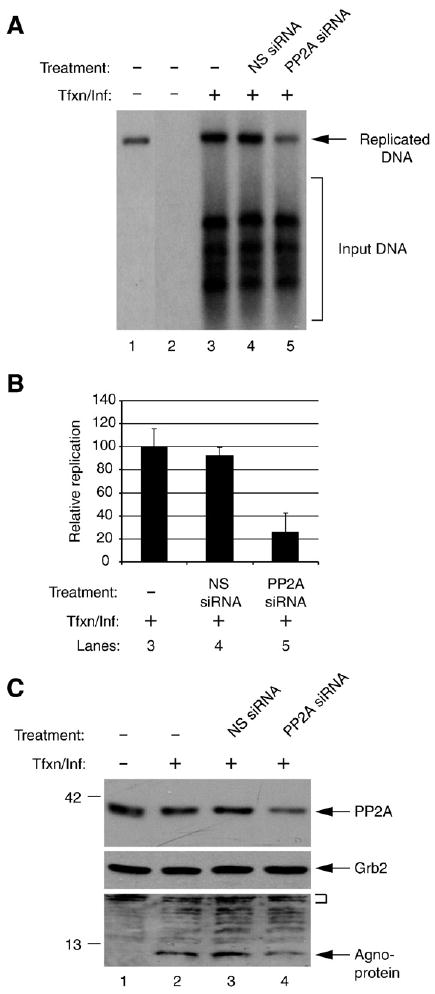
Effect of PP2A inhibition on JCV replication and gene expression. (A) Inhibition of PP2A negatively affects JCV replication. SVG-A cells. 2 × 105 cells per 35-mm tissue culture dish were transfected/infected with JCV genome (2 μg/plate) in the presence and absence of 50 pmol of non-targeting siRNA or Smartpool PP2AC siRNA (PP2AC siRNA, Dharmacon, Lafayette, CO). At the 48 h post infection, cells were trypsinized and split into two equal portions. One half of the samples were used to prepare low molecular weight DNA for Dpn I assay and the other portion was used to prepare whole cell extracts for Western blotting. DNA was digested with both Bam HI and Dpn I and analyzed by Southern blotting using labeled JCV genomic DNA as probe. Dpn I enzyme digests only transfected (input) DNA and leaves intact the newly replicated DNA. In lane 1, plasmid containing the Mad-1 genome was digested with Bam HI only and 0.1 ng of it was run on the gel as a positive control. In lane 2, the DNA sample from untransfected/uninfected cells were processed as in lanes 3–5 and was loaded as a negative control. The arrow points to the replicated DNA and the bracket indicates the input DNA. Tfxn/Inf denotes transfection/infection, i.e., introduction of the Mad-1 JCV genome into cells by transfection. (B) Quantitative representation of the results from panel A. The bands corresponding to the replicated DNA was quantitated in each lane (lanes 3–5) by densitometry and normalized relative to the band intensity on lane 3. The standard deviations of the data are presented as error bars (C) Western blot analysis of the whole cell extracts prepared in parallel to the replication assay in panel A, using specific antibodies against agnoprotein, PP2AC and Grb2. In lane 1, sample from untransfected/uninfected cells was loaded as a negative control. The small bracket denotes the non-specific bands that indicate the equal protein loading for agnoprotein. Grb2 was probed as a loading control for PP2AC.
