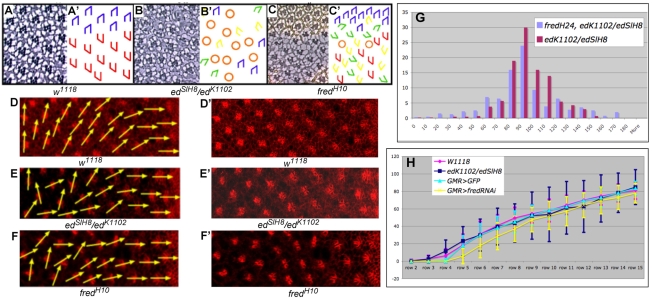
Fig. 1.
ed and fred mutant ommatidia misrotate. (A-C′) Tangential sections through adult Drosophila eyes (A-C) and corresponding schematics (A′-C′). (A) Wild type (w1118). Ommatidia come in two chiral forms, depicted in blue in the dorsal half and red in the ventral half of the eye. (B) Some edSlH8/edK1102 ommatidia under- or over-rotate (green and yellow trapezoids, respectively), and some contain an incorrect number of photoreceptors (orange circles). (C) Some ommatidia in fredH10 clones rotate correctly whereas others under- or over-rotate. (D-F′) The ed and fred orientation phenotypes result from aberrant ommatidial rotation. Anti-Arm staining (red) highlights the apical surface and outlines cell boundaries. Yellow vectors bisect R8 and run through the R3/R4 interface, highlighting the angle of orientation of each ommatidium. (D) Wild-type ommatidia follow a smooth progression of rotation. Ommatidial precursors in both edK1102/edSlH8 (E) and GMR>fredRNAi (F) knockdown eye discs misrotate. D′-F′ show D-F without arrows. (G) Reduction of fred activity enhances the ed mutant phenotype. Bar chart illustrating the percentage of ommatidia (y-axis) that are oriented at the angles indicated (x-axis) in edk1102/edSlH8 and edK1102, fredH24/edSlH8 eyes. (H) Graphical representation of data from D-F plotted as the mean angle of orientation (MAO) of ommatidia in each of four genotypes in rows 2-15. Error bars indicate the variance (s.d.). w1118 is the control for edK1102/edSlH8; GMR>GFP is the control for GMR>fredRNAi. The s.d. of ed and fred ommatidia is significantly different from that of the controls between rows 7-15. Trapezoid color for all schematics: blue and red, wild-type; green, under-rotated; yellow, over-rotated; black, fail to rotate; orange circles, incorrect number of photoreceptors.