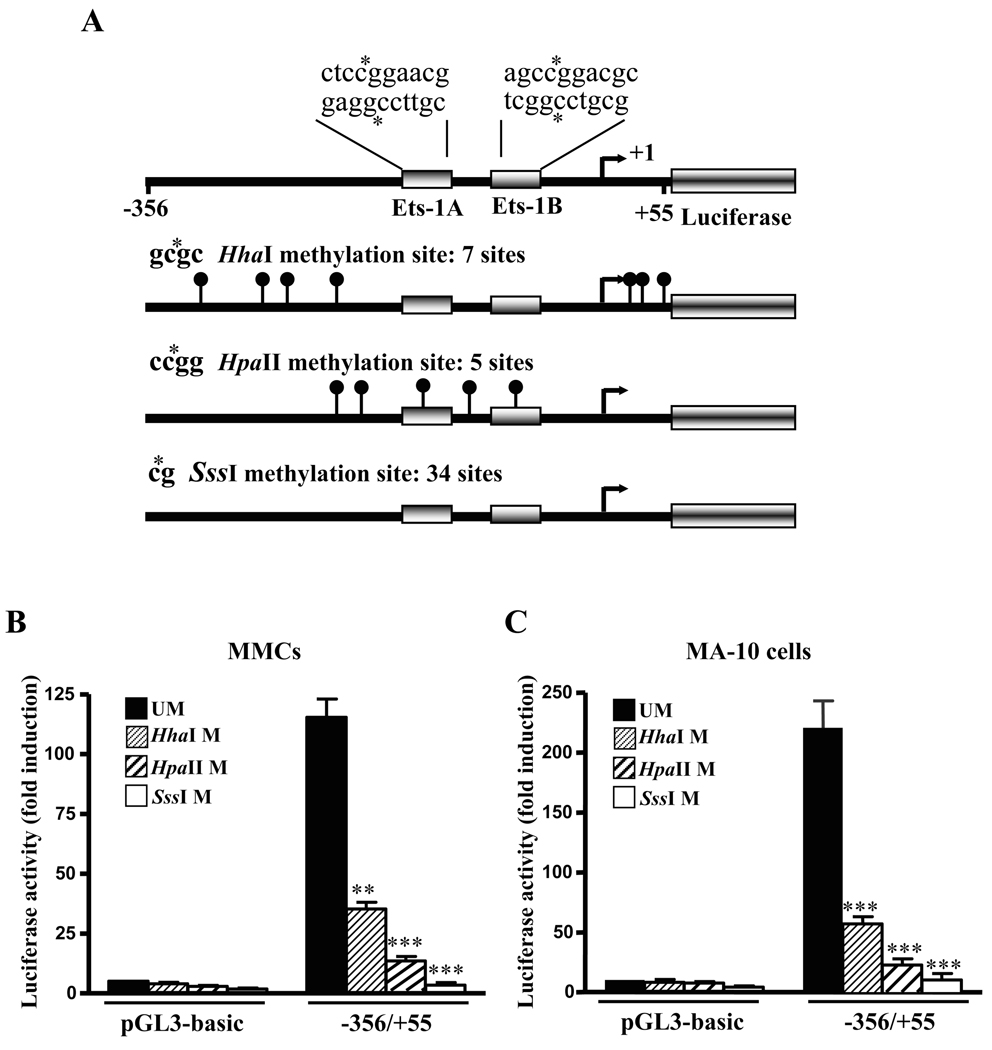
Fig. 7. Effect of methylation on Npr1 promoter activity.
A) Schematic representation of the potential CpG methylation island of −356/+55 in Npr1 promoter. The construct contains 411 bp between positions −356 to +55 relative to the TSS (bent arrow), which is indicated as +1. Boxes represent Ets-1A and Ets-1B binding sites. Location of methylation sites by HhaI, HpaII and SssI methylases are indicated as lollipop, together with the recognition sequences of the methylases. Asterisk indicates the position of methylated cytosine by each methylase. Luciferase activity of the in vitro methylated pGL3-basic and −356/+55 Npr1 promoter construct transfected in (B) MMCs and (C) MA-10 cells. The luciferase activity of the constructs was compared with those of corresponding unmethylated constructs. UM indicates unmethylated; M indicates methylase. Bars represent the mean ± SE of four independent experiments in triplicate. **, p < 0.01, ***, p < 0.001 versus unmethylated construct.