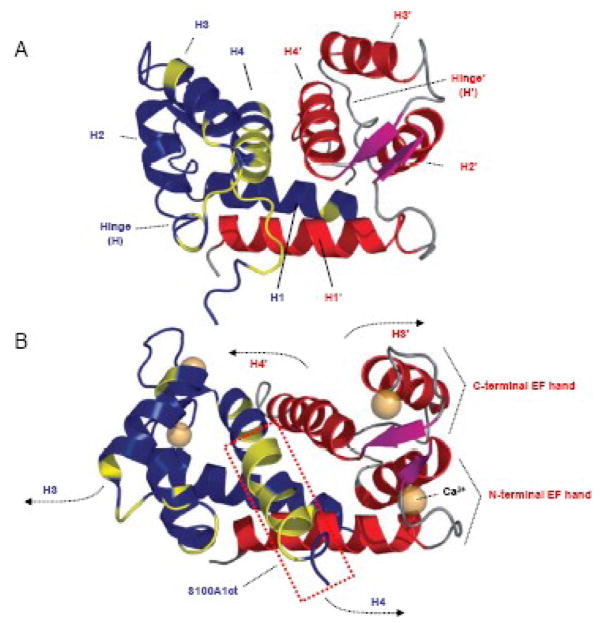
Figure 2. Three dimensional (3D) solution structure of S100A1 determined by NMR spectroscopy.
A,3D S100A1 structure in the apo state: S100A1 is composed of two identical subunits (blue/red) each composed of a repetitive helix-loop-helix calcium binding motif that is connected by a short linear linker (hinge) region. Dimerization occurs in an antiparallel manner between helix (H) 1 and 1′ stabilized by hydrophobic bonds. Hydrophobic residues in the hinge region and H4 in the blue coloured dimer are shown in yellow. This 3D structure reflects the characteristic molecular backbone of S100 proteins. B, 3D S100A1 structure in the calcium bound state: Ca2+ binding both to the N- and C- terminal EF hand motif mainly results in altered orientation of H3 and H4 as well as the hinge region while the residual structure of the dimer remains nearly unaffected. As indicated by arrows, the N-terminal part of H3/3′ and the C-terminal part of H4/4′ in both dimers swing apart exposing hydrophobic residues of both helices and the hinge region to the surface facilitating their Ca2+ dependent interaction with target molecules. S100A1ct is indicated by the dashed red box. Reproduced with modifications from Wright et al. [27].