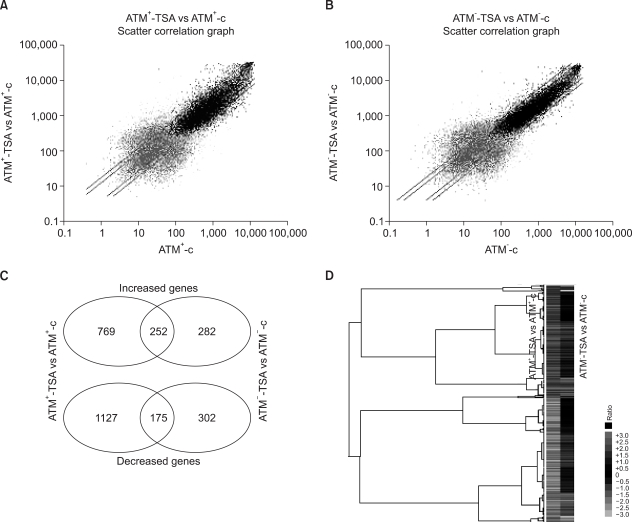
Fig. 1.
Comparison of the gene expression profiles between the ATM- and ATM+ cells in response to TSA, as was derived from the oligonucleotide microarray analysis. (A) The signal intensities from the TSA-treated ATM+ cells (y-axis) relative to those from the untreated ATM+ cells (x-axis) were analyzed using MicroArray Suite 5.0 software. The log-transformed data is shown; the guideline shows the 2-fold signal ratio. (B) The signal intensities in the ATM- cells. The ATM-dependent and ATM-independent TSA-responsive genes were distinguished from the Venn diagrams (C), and the genes that were significantly enhanced or reduced in the ATM+ cells, as compared to that in the ATM- cells, in response to TSA were visualized by Treeview software (D). The expressions of more genes are regulated in the ATM+ cells, as compared to the ATM- cells, in response to HDAC inhibition.