Abstract
Acute pancreatitis is a serious and potentially fatal disease caused by intracellular trypsinogen activation. Although protease detection has been greatly facilitated by the development of protease probes capable of monitoring protease activation and inhibition, real-time quantitative measurement of protease activity in living cells remains a challenge, and the identification of the cellular compartment for trypsinogen activation is inconclusive. Here we report a novel strategy for developing trypsin sensors by grafting an enzymatic cleavage site into a sensitive location for optical change of chromophore in a single enhanced green fluorescent protein (EGFP). Our designed trypsin sensor exhibits rapid kinetic responses for protease activation and inhibition with a large ratiometric optical signal change. In addition, it has strong specificity, as enzymatic cleavage is not observed with other proteases such as thrombin, cathepsin B, tryptase, and tissue plasminogen activator. Moreover, the developed trypsin sensor allows us for the first time to observe, in real time, trypsinogen activation by caerulein in the pancreatic cancer cell line, MIA PaCa-2 without zymogen granules. These developed protease sensors will facilitate improved understanding of mechanisms and locations of protease activation and further provide screening of protease inhibitors with therapeutic effects.
Keywords: EGFP, protease sensors, trypsinogen activation, ratiometric measurement, pancreatic cancer cell, granule, protease inhibitor
Digestive proteases are produced as inactive proenzymes inside the membrane-bound zymogen granules (1). The proenzyme becomes active after physiological stimulation during exocytosis (2, 3). Usually protease inhibitors coexist with proteases at concentrations sufficient to prevent autodigestion (4). Fatal human diseases such as acute pancreatitis result from autodegradation of trypsin, chymotrypsin and elastase derived from pancreatic tissues. Trypsin is one of the major digestive enzymes responsible for regulating protein degradation (4). Misbalanced trypsinogen secretion, activation, inhibition, and turnover can result in acute and chronic pancreatic diseases (5). Approximately 35,000 new cases of pancreatic cancer associated with misbalanced serine protease activity, especially trypsin, are reported annually. It is the fourth leading cause of cancer deaths in the United States (6). Diagnosis of chronic pancreatic diseases is largely restricted to later stages of the illness, partly due to inherent limitations in available peptide kits designed for the detection of trypsin activity in cell lysates. Caerulein is the most commonly used inducer of trypsin activation in living pancreatic acinar cells (1, 7). It is widely accepted that zymogen is packed into granules of acinar cells. However, the location of trypsinogen activation has not been conclusively identified despite substantial studies using various methods. Several studies have suggested that zymogen activation might be initiated in zymogen granules (2, 8). Using antibodies against trypsinogen activation peptide in confocal and immunoelectronic microscopy, initial zymogen activation was observed in a vesicular non-zymogen granule compartment (9, 10). The activation of trypsinogen in large endocytic vacuoles has also been proposed to initiate acute pancreatitis (3).
Currently, the pancreatic cancer cell line, MIA PaCa-2 derived from human pancreatic tumor tissue (11), has been extensively used in developing cancer therapy based on protease-induced apoptosis (12, 13). Differing from acinar cells, pancreatic cancer cells such as MIA PaCa-2 do not have a granule structure (14). However, an elevated mRNA level of trypsinogen and secretory trypsin inhibitor was reported in this cancer cell line (15, 16). To date, it is still unknown whether pancreatic cancer cells such as MIA PaCa-2 have any elevation of trypsin activity. It is also not clear, due to limitations of current commercially available trypsin probes, whether trypsinogen activation of pancreatic cells occurs in the cytosol.
Stimulated by our previously-developed calcium sensors based on a single EGFP (17), here we report the development of a trypsin sensor by engineering an enzymatic cleavage site into a single wild type EGFP (EGFP-wt) (Figure 1a and 1b). We have shown that the developed trypsin sensor exhibits large ratiometric fluorescence signal change upon cleavage at the designed cleavage site in EGFP. More importantly, the developed trypsin sensor exhibits strong enzymatic selectivity over other proteases to permit real-time monitoring of protease activity in living cells. Moreover, we have shown that trypsinogen activation also can be triggered by caerulein in MIA PaCa-2 cancer cells.
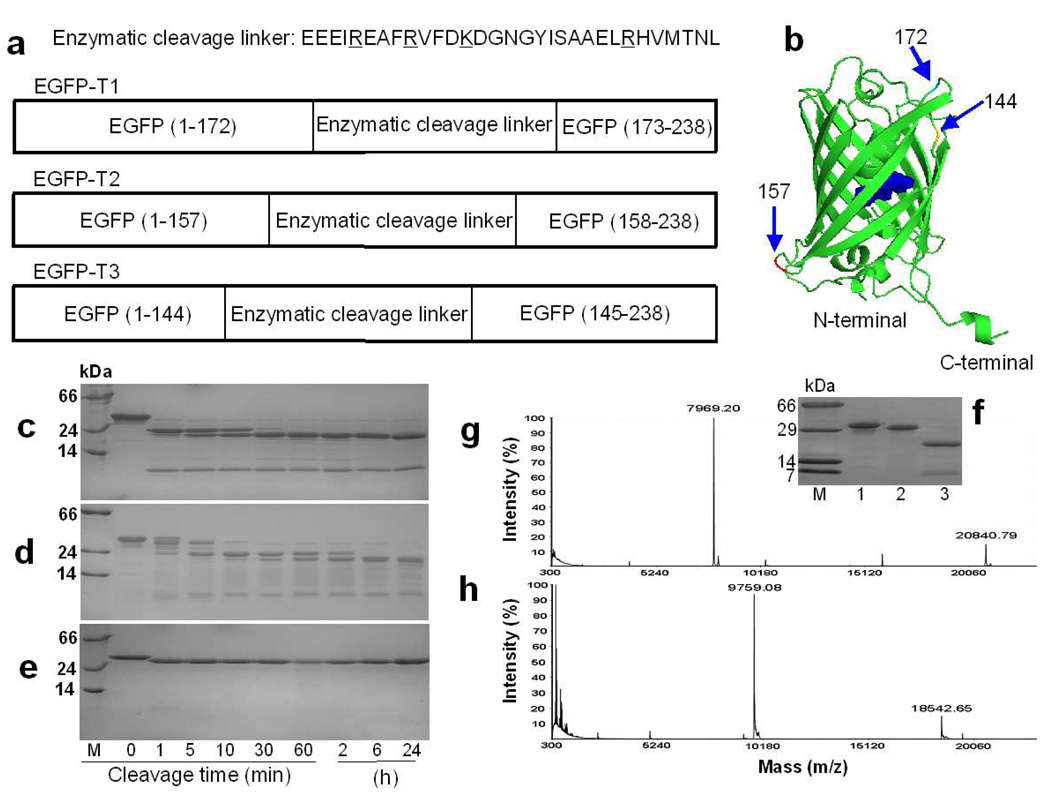
Figure 1.
The design and cleavage confirmation of trypsin sensors. a) Three EGFP-based trypsin sensors with grafted cleavable linkers at position 144, 157 and 172. Trypsin cleavage occurs following residues R or K (underlined in sequence). b) Insertion locations for trypsin linker in EGFP. SDSPAGE gels show the cleavage products of EGFP-T1 (c), EGFP-T2 (d), and EGFP-wt (e) at different digestion time intervals. M is the protein marker. Lane 1, 2, 3, 4, 5, 6, 7, 8 and 9 are samples following trypsin digestion for 0 min, 1 min, 5 min, 10 min, 30 min, 60 min, 2 h, 6 h, and 24 h at room temperature, respectively. f) To block the trypsin cleavage site, all Arg and Lys in the cleavage linker of EGFP-T1 were mutated into Ala to obtain EGFP-T1m. This mutation prevented cleavage in the inserted linker of EGFP-T1m. M is the protein marker. Lane 1, 2 and 3 are samples of EGFP-T1m without tryspin digestion, EGFP-T1m with trypsin digestion and EGFP-T1 with trypsin digestion, respectively. The product fragments of EGFP-T1 (g) and EGFP-T2 (h) cleaved by trypsin were identified by MALDI-MS.
Experimental Section
Construction of trypsin sensors
A cleavage linker (EEEIREAFRVFDKDGNGYISAAELRHVMTNL) for trypsin was genetically generated in Pet28a plasmid (Invitrogen) encoding EGFP at three locations (Glu172, Gln157 and Asn144) using polymerase chain reaction (PCR) techniques. The resulting variants of trypsin sensors were labeled EGFP-T1, EGFP-T2 and EGFP-T3, respectively. A modified linker (EEEIAEAFAVFDADGNGYISAAELAHVMTNL) without cleavage sites for trypsin and a modified cleavage linker (EEEIAEAFAVFDADGNGYISAGPARHVMTNL) without calcium-binding capability were also obtained through PCR amplification and labeled EGFP-T1m and EGFP-T1nb, respectively. Different EGFP-based trypsin sensors were then sub-cloned into pcDNA3.1(+) (Invitrogen) for mammalian expression.
Spectral properties following trypsin digestion
Trypsin digestion of 15 µM EGFP-T1 and EGFP-T2 was performed by adding stock trypsin from bovine pancreas (Sigma, St. Louis) up to a final concentration of 20 nM in trypsin reaction buffer (10 mM Tris, 20 mM CaCl2, pH 7.4). The UV-visible spectra following trypsin digestion were measured in a 1 cm pathlength cuvette at different time intervals (0, 1, 3, 5, 10, 20, 30, 60, 90 and 120 min) and fluorescence spectra of trypsin sensors with excitation at 398 and 490 nm were recorded at different time intervals (0, 1, 3, 5, 10, 20, 30, 60, 90 and 120 min). Similarly, EGFP-wt was subjected to identical digestion as the control.
Cleavage status verification
To examine the cleavage status of trypsin sensors at different times following trypsin digestion, 20 µL samples of 15 µM EGFP-T1, EGFP-T2 and EGFP-wt were removed, immediately combined with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and boiled to denature the proteins and halt the digestion reaction for SDS-PAGE analysis. Samples following digestion were evaluated at 0 min, 5 min, 10 min, 30 min, 60 min, 2 h, 6 h and 24 h, respectively. Additionally, the cleavage products of trypsin sensors following trypsin digestion for 24 hours were also collected and analyzed using matrix assisted laser desorption/ionization-mass spectrometry (MALDI-MS).
Kinetic studies
Initial rates of trypsin digestion reaction for EGFP-T1 at various concentrations were measured using a time-course model to monitor the change of absorbance at 490 nm for 10 min on a UV-1700 spectrometer (Shimadzu, Japan), where the initial rate was equivalent to the resulting slope of absorbance increase. These data were then used to calculate the steady-state kinetic parameters, Michaelis constants (Km), turnover number (kcat) and second order rate constants (kcat/Km) for hydrolysis of EGFP-T1 following trypsin cleavage, by fitting data to the Michaelis-Menten equation (Eq.1) by nonlinear regression using KaleidaGraph 3.5 software (Synergy software).
| Eq. 1 |
In Equation 1, v0/e is per mole enzyme initial reaction rate (s−1), kcat is the turnover number (s−1), [S] is substrate concentration (M), and Km is the Michaelis constant (M).
Cell culture and imaging
MIA PaCa-2 cells were plated on sterilized glass coverslips in 35-mm dishes and grown to 60% confluence in DMEM (Sigma Chemical Co., St. Louis, MO) supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37 °C with 5% CO2 in a humidified incubation chamber. Cells transfected with EGFP-T1, EGFP-wt and EGFP-T1m were allowed to grow for 24–48 h before imaging (17, 18). In addition, 1 µM soybean trypsin inhibitor (SBTI) was incubated with cells transfected with EGFP-T1 for 30 min before imaging. EGFP-wt, EGFP-T1m and EGFP-T1 with SBTI inhibition were used as the controls. Cells were imaged on a Zeiss Axiovert 200 Inverted Microscope with a 40x oil objective lens and a CCD camera (AxioCam HRc). Ratiometric emission imaging was acquired through an FITC filter set (excitation wavelength: 480 ± 20 nm; emission wavelength: 510 ± 20 nm) and 395 nm filter set (excitation wavelength: 395 ± 20 nm; emission wavelength: 510 ± 20 nm) from a light source (FluoArc, Zeiss) in a time-course mode using Axiovision software. During image acquisition, caerulein was added to the cell plate to a final concentration of 10 nM for trypsin activation (19). The ratiometric change between fluorescence emission for excitation at 488 and 398 nm was calculated at different time intervals to detect trypsin activity in living cells. All fluorescence emission at different time intervals was normalized by dividing initial fluorescence intensity to set the basal ratio to 1 (20).
The fluorescence ratiometric change (R) is expressed by equation 2:
| Eq. 2 |
where NFt-488 is the normalization fluorescence intensity of various time points at 488 nm excitation and NFt-398 is the normalization fluorescence intensity of various time points at 398 nm excitation.
Trypsin activation assays in cell lysates
To examine trypsin activity and confirm trypsin activation pattern in living cells, MIA PaCa-2 cells (1 × 106) were cultured in 100-mm cell culture dishes for 24 h and then treated with 10 nM caerulein. The cell pellets following caerulein induction at various timeintervals (0, 10, 20, 30, 60, 120, 240 min and 24 h) were harvested and lysed with lysis buffer (10 Mm Tris, 10 mM NaH2PO4/NaHPO4, 150 mM NaCl, 0.1% Triton X-100, 10 mM Na2P2O7·10H2O, pH 7.5). The supernatant containing protein was obtained through centrifugation at 13000×g. The total protein concentration was determined using the BCA assay kit (Pierce, USA) according to the protocol from the manufacturer. The cell lysates containing 20 µg protein of each sample at various time intervals were mixed with 100 µl of 4 mM trypsin substrate kit, Boc-Gln-Ala-Arg-AMC, in a total volume of 250 µl of trypsin reaction buffer (10 mM Tris, 20 mM CaCl2, pH 7.4), and then incubated under light-free conditions for 30 min at 37 °C. Following incubation of the reaction mixture, fluorescence intensity at 450 nm of AMC released from trypsin substrate kit due to the cleavage by active trypsin was measured using a fluorescence microplate reader under excitation at 380 nm. The relative fluorescence units (RFU) in the induced samples relative to control buffer sample indicate the relative amount of trypsin. Comparing RFU of the positive control, active trypsin, the amount of trypsinogen activation can be estimated at various time points following caerulein stimulation in living cells.
Results and Discussion
We hypothesized that the proteolytic cleavage occurring within designed sequences at sensitive locations near the chromophore could result in a chromophore signal change due to local conformational change. Three grafting sites on the EGFP scaffold were selected based on the capability of the chromophore signal change to monitor dynamic enzymatic activity in living cells. Three criteria were identified during the design of the sensors as being critical for development. First, protease sensors must be able to dynamically monitor enzymatic activation or inhibition in intact living cells in real time. Second, the optical signal change of sensors should have good selectivity, high sensitivity and rapid response to catalytic cleavage with an optimal kinetic activity during protease cleavage. For trypsinogen activation, it is important to have enzymatic selectivity against other proteases such as cathepsin B and tryptase that are well known to be involved in the trypsinogen activation (9, 21, 22). Third, to measure enzymatic activity quantitatively, the change of fluorescence emission or excitation signals induced by proteases should be ratiometric to eliminate problems associated with variations in sensor concentrations, subcellular conditions, instrument instability, and photobleaching (20, 23). Fourth, a large dynamic signal change following enzymatic cleavage allows for the rapid and early detection of protease activation and sensitive screening of therapeutic inhibitors as drugs. Since trypsin demonstrates a strong preference to cleave the peptide bond immediately following an Arg or Lys residue in a solventaccessible region, a trypsin cleavage sequence derived from EF-hand motif loop III of calmodulin was selected as the basic graft structure because it contains highly-accessible Arg and Lys residues in the loop and flanking helices. As shown in Figure 1b, the selected sites appear in the sequence immediately following Glu172 within loop-9, Gln157 within loop-8, and Asn144 within loop-7. The resulting sensors were designated EGFP-T1, EGFP-T2 and EGFP-T3, respectively. To eliminate the contribution of calcium binding, Glu at the EF-loop position 12 was replaced by Ala to obtain EGFP-T1nb variant with a single tryspin cleavage site (GPAR sequence at positions P4-P1 around the cleavage site).
The engineered protease sensors along with EGFP-wt were expressed in E. coli and purified with a chelating affinity column. To verify the cleavage status of EGFP-based trypsin sensors, each sensor was incubated with trypsin at various digestion time intervals and the resulting cleavage products were examined using SDS-PAGE and MALDI-MS. The results indicated that EGFP-T1 and EGFP-T2 were specifically cleaved into two major fragments by trypsin at the grafted cleavage site, approximately 20 and 8 kDa (Figure 1c) from EGFP-T1 and 18 and 10 kDa (Figure 1d) from EGFP-T2. In contrast, EGFP-wt exhibited only a minor decrease in mass (Figure 1e) resulting from the removal of His-tag upon incubation with trypsin. Furthermore, we created a negative control EGFP variant (EGFP-T1m) through mutation of Arg and Lys within the designed cleavage linker to Ala. Upon addition of trypsin, EGFP-T1m did not appear to be cleaved into small fragments at the inserted linker region in SDS-PAGE (Figure 1f) although a similar minor decrease in mass of EGFP-wt was observed due to the removal of His-tag. The absorption and fluorescence signals of EGFP-T1m are also not altered upon addition of trypsin. These results clearly demonstrate that cleavage occurs specifically at the designed cleavage linker in EGFP-T1 and EGFP-T2.
These two major trypsin cleavage fragments from EGFP-T1 remained unchanged until digestion time exceeded 40 hours. MALDI-MS analysis revealed that fragments produced from cleavage of EGFP-T1 had molecular masses of 20840.79 and 7969.20 Da (Figure 1g), and fragments from cleavage of EGFP-T2 had molecular masses of 18542.65 and 9759.08 Da (Figure 1h), respectively, which is consistent with cleavage occurring in the cleavage linker. Conversely, the non-fluorescing variant EGFP-T3 was not suitable as a sensor due to its apparent unfolded structure as revealed by far UV circular dichroism (data not shown). In order to further verify the accurate cleavage location, both fragments were separated and subjected to Edman degradation protein sequencing. Based on the comprehensive analyses of SDSPAGE, MALDI-MS and N-terminal protein sequencing, a clear cleavage pattern of EGFP-T1 following trypsin digestion was determined to reveal the removal of His-tag and specific cleavage in the inserted linker. The sequence of the large fragment is from MVSKGEE to EAFR inside the linker. The sequence of the small fragment is from HVMTNL to the C-terminal of EGFP-T1.
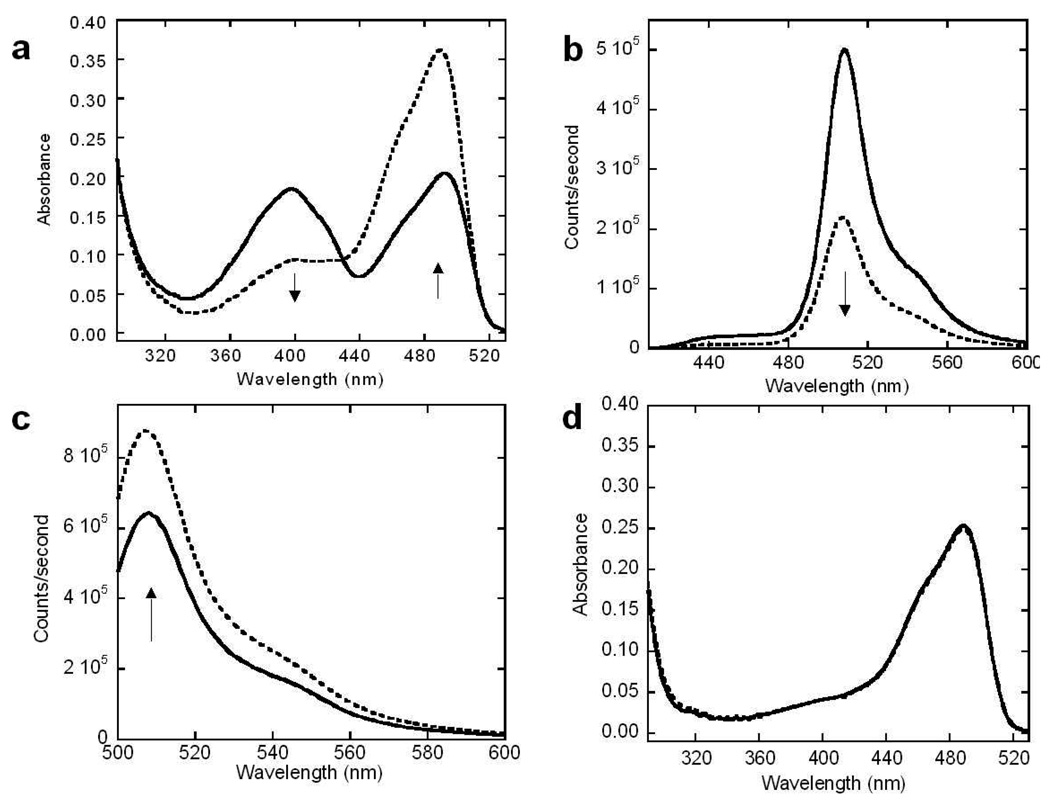
EGFP-T1 and EGFP-T2 retained strong fluorescence both in bacteria and as purified proteins. EGFP-T2 has a maximum absorption wavelength similar to that of EGFP-wt at 490 nm originating from the deprotonated form of the chromophore, while EGFP-T1 has two strong absorption peaks at 490 and 398 nm with extinction coefficient constants of 16235 and 14137 M-1cm-1, respectively (Supporting Figure S1a), where the peak at 398 nm corresponds to the protonated chromophore species.(24) Moreover, the maximum emission of EGFP-T1 at 508 nm with a quantum yield of 0.59 results from excitation at 490 nm (Supporting Figure S1b). Following trypsin digestion, the maximum absorption of EGFP-T1 at 490 nm increased and concurrently decreased at 398 nm (Figure 2a). Similarly, EGFP-T1 exhibited a ratiometric signal change with a decrease in maximum fluorescence emission intensity at 508 nm at an excitation wavelength of 398 nm (Figure 2b) and a corresponding increase at an excitation wavelength of 490 nm (Figure 2c). In contrast, EGFP-wt, which was not cleaved, exhibited no optical signal change and the maximum absorption of EGFP-T2 showed no obvious change although it was cleaved by trypsin under identical digestion conditions (Figure 2d). This result suggests that position 172 in EGFP is a sensitive location where the ionic state of the chromophore species can be altered by the addition of the grafting sequence. The dual chromophore forms in GFP have also been demonstrated in previous reports (24). A complicated hydrogen bond network allowing for proton transfer between the chromophore and its neighboring side chains significantly contributes to the equilibrium between protonated and deprotonated chromophore forms (25). The predominant protonated form of the chromophore exhibits its maximum absorbance at 397 nm, which is governed by the carboxylation of Glu222 through electrostatic repulsion and hydrogen bonding between a bound water molecule and Ser205. In contrast, the maximum absorbance peak of the deprotonated form of the chromophore is observed at 490 nm. Under this condition, the charge of Glu222 was proposed to donate to the chromophore through proton abstraction and a hydrogen bond network, involving Ser205 and bound water. In order to stabilize the deprotonated state, a rearrangement of the side chains of Thr203 and His148 may occur. In addition to the proton relay between Ser205 and Glu222, the side chains of Asp117, Thr118, Glu172 and Asp190 were also proposed to be involved in the alteration of different conformations of the chromophore, as observed in the electron density of the crystal structure (25). Our observation of the switch of dual chromophore forms following the insertion of cleavage linker at position 172 is likely due to alteration of the ionization state of the chromophore. Following the insertion of the cleavage linker at position 172 of EGFP, the population of both states of the chromophore may be significantly altered through the proton relay between Ser205 and Glu222 and rearrangement of hydrogen bond networks due to the environmental change of Glu172 (25). The observed changes in maximum absorption and fluorescence emission intensities were due to changes in relative proportions of the two chromophore species of EGFP-T1 following trypsin digestion. The dynamic range of EGFP-T1 sensor is 3.8 for fluorescence, which is comparable to current protein-based sensors (26-29).
Figure 2.
Optical change and kinetic studies of trypsin sensors. Absorption spectra of EGFP-T1 (a) were measured after trypsin digestion at various times from 0 min (solid line) to 120 min (dashed line) in trypsin digestion buffer (10 mM Tris, 20 mM CaCl2, at pH 7.4). Absorbance of EGFP-T1 decreased at 397 nm and increased at 491 nm following trypsin digestion at different digestion time intervals. The fluorescence spectra of EGFP-T1 with excitation at 398 nm (b) and excitation at 490 nm (c) were measured following trypsin digestion from 0 min (solid line) to 120 min (dashed line) in trypsin digestion buffer, respectively. Fluorescence of EGFP-T1 decreased when excited at 397 nm and increased when excited at 491 nm following trypsin digestion at different digestion time intervals. There is not an absorption spectrum change of EGFP-T2 (d) following a trypsin digestion.
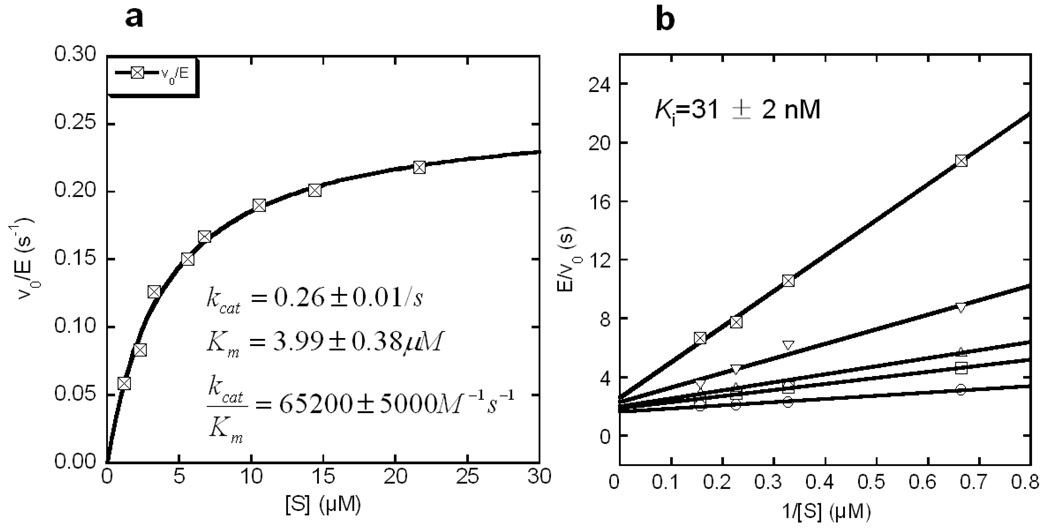
Kinetic studies of EGFP-T1 were compared with results from three commercially available trypsin kits, Bz-DL-Arg-pNA (BAPNA), Bz-DL-Arg-AMC and Boc-Gln-Ala-Arg-AMC under identical buffer conditions. All of these commercially available kits exhibit optical signal changes at a single wavelength (i.e. non-ratiometric) upon trypsin cleavage. Kinetic parameters, kcat, Km and kcat/Km of EGFP-T1 (Figure 3a) and the commercial trypsin kits were calculated through fitting with Eq(1). Kinetic parameters are shown in Table 1. The EGFP-T1 sensor exhibited a 750-fold decrease in Km and a 150-fold increase in kcat/Km compared to the BAPNA colorimetric kit. Similarly, an approximately 20-fold decrease in Km and a 20-fold increase in kcat/Km were observed in comparison to Bz-DL-Arg-AMC. Boc-Gln-Ala-Arg-AMC with preferable residues at positions P2 and P3 exhibited 2.3-fold larger Km and 5.4- fold smaller kcat than EGFP-T1. Consequently, trypsin cleavage of EGFP-T1nb occurred at the designed cleavage site and kcat/Km of EGFP-T1nb for trypsin increased relative to that of Boc-Gln-Ala-Arg-AMC due to the addition of preferable residues for trypsin at P2 and P3 positions before the trypsin cleavage site. Overall, kcat/Km of EGFP-based trypsin sensor variants is comparable to that of Boc-Gln-Ala-Arg- AMC.
Figure 3.
Kinetic studies of EGFP-T1 were conducted in trypsin digestion buffer. The kinetic parameters, kcat, Km, and kcat/Km, were determined to be 0.26 ± 0.01/s, 3.99 ± 0.38 µM, and 65200 ± 5000 M−1s−1 through Michaelis-Menten equation fitting (a), respectively. EGFP-T1 with trypsin inhibitor, leupeptin with 0 nM (○), 12.5 nM (□), 25 nM (△), 50 nM (▽), 100 nM (⊠) is evaluated (b) to reveal the competitive inhibition to trypsin, with Ki value of 31 ± 2 nM.
Table 1.
Comparison of kinetic parameters between our trypsin sensors and commercial trypsin kits
| Trypsin sensors or kits | kcat(s−1) | Km(µM) | kcat/Km (M−1s−1) |
|---|---|---|---|
| EGFP-T1 | 0.26 ± 0.01 | 3.99 ± 0.38 | (6.52 ± 0.50) × 104 |
| EGFP-T1nb | 0.85 ± 0.06 | 4.21 ± 0.71 | (2.01 ± 0.21) × 105 |
| Bz-DL-Arg-pNA | 1.33 ± 0.17 | (3.00 ± 0.50) × 103 | (4.44 ± 0.41) × 102 |
| Bz-DL-Arg-AMC | 0.25 ± 0.01 | 77.44 ± 8.46 | (3.22 ± 0.21) × 103 |
| Boc-Gln-Ala-Arg-AMC | 1.41 ± 0.05 | 9.27 ± 0.95 | (1.52 ± 0.12) × 105 |
In order to ensure that the developed protease sensor is able to directly monitor the catalytic step, the inhibition effect of leupeptin on trypsin was examined during the digestion of EGFP-T1 with various leupeptin concentrations. The rate of the signal change due to cleavage was found to decrease with increasing leupeptin concentrations (Supporting Figure S2). The rates of EGFP-T1 with various leupeptin concentrations were fitted to reveal a competitive inhibition with a Ki value of 31 ± 2 Nm (Figure 3b). This clearly demonstrates that the optical signal change of the EGFP-T1 sensor is directly related to enzymatic cleavage rather than the binding process and can be applied to monitor inhibition processes for drug screening.
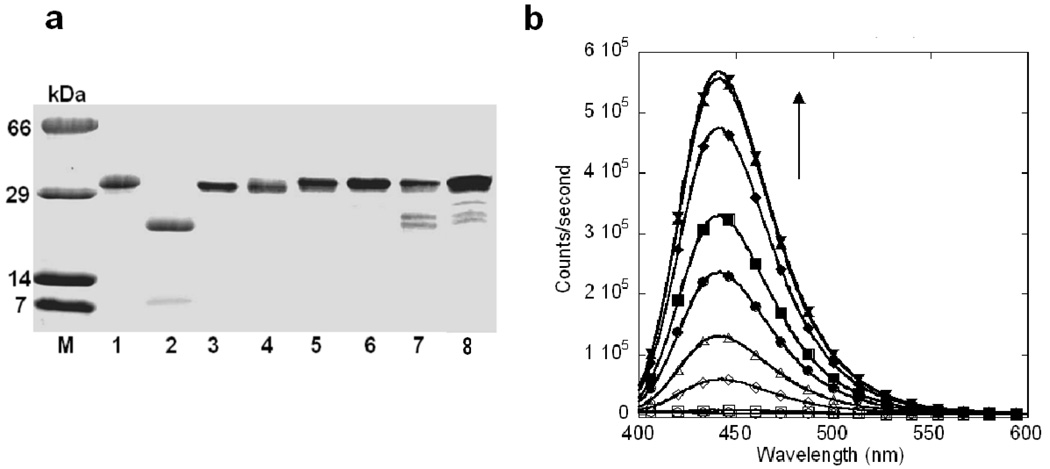
One of the major challenges in developing protease probes is to achieve strong specificity for different proteases (30). To examine the enzymatic specificity of EGFP-T1, thrombin, cathepsin B, tryptase, tissue plasminogen activator, kallikrein and chymotrypsin from mammalian proteinase family with different substrate specificities were incubated with this sensor. Lysosomal cathepsin B was shown to be responsible for the trypsinogen activation (9, 21, 22). Cathepsin B, tryptase, thrombin and tissue plasminogen activator could not cleave EGFP-T1 as monitored by SDS-PAGE (Figure 4a). Less than 10–30% of EGFP-T1 was cleaved by kallikerin and chymotrypsin upon overnight incubation of cleavage at the same concentration of trypsin, suggesting that the designed protease sensor with a specific cleavage linker exhibited strong enzymatic specificity. In contrast, cathepsin B substrate, Z-Phe-Arg- AMC, was cleaved under identical buffer conditions in 5 min by cathepsin B, as indicated by the fluorescence signal change (Figure 4b). Such strong enzymatic selectivity is very exciting since cathepsin B and tryptase are known to cleave the residues after Arg, which are involved in trypsinogen activation. Thus, our developed sensor is ideal for cellular applications to understand the mechanisms of trypsinogen activation.
Figure 4.
The selectivity of EGFP-T1 for proteases. SDS-PAGE (a) exhibited no cleavage bands of EGFP-T1 following 20 nM thrombin, cathepsin B, tryptase or tissue plasminogen activator overnight digestion, but EGFP-T1 can be slowly cleaved by kallikerin and chymotrypsin. M is protein marker. Lane 1 is EGFP-T1 without digestion. Lanes 2, 3, 4, 5, 6, 7 and 8 are EGFP-T1 following overnight digestion with trypsin, thrombin, cathepsin B, tryptase, tissue plasminogen activator, kallikerin and chymotrypsin, respectively. To confirm optimal reaction buffer for EGFP-T1 cathepsin B digestion, ZPhe- Arg-AMC for cathepsin B (b) was cleaved under identical digestion buffer conditions to reveal fluorescence signal increase following digestion time increase at 0 (○), 1 (□), 5 (◊), 10 (△), 20 (●), 30 (■), 60 (◆), 120 (▲) and 180 min (▼) through detection by fluorescence spectroscopy.
To further test the pH effect on our trypsin sensor, we have also conducted experiments to examine the cleavage status of our EGFP-T1 in buffers at various pH values (pH 4–9). As shown in Supporting Figure S3, trypsin sensor EGFP-T1 can be cleaved by trypsin under buffer conditions from pH 5 to Ph 9. In addition, we have conducted a kinetic study of trypsin sensor (EGFP-T1) to examine the effect of pH on the kinetic parameters. Less than two-fold increase in kcat/Km value for EGFP-T1 was observed when the buffer condition changed from pH 6.7 to 8.0, suggesting that the developed trypsin sensor is able to specifically respond to the trypsin cleavage from pH 5 to 9.
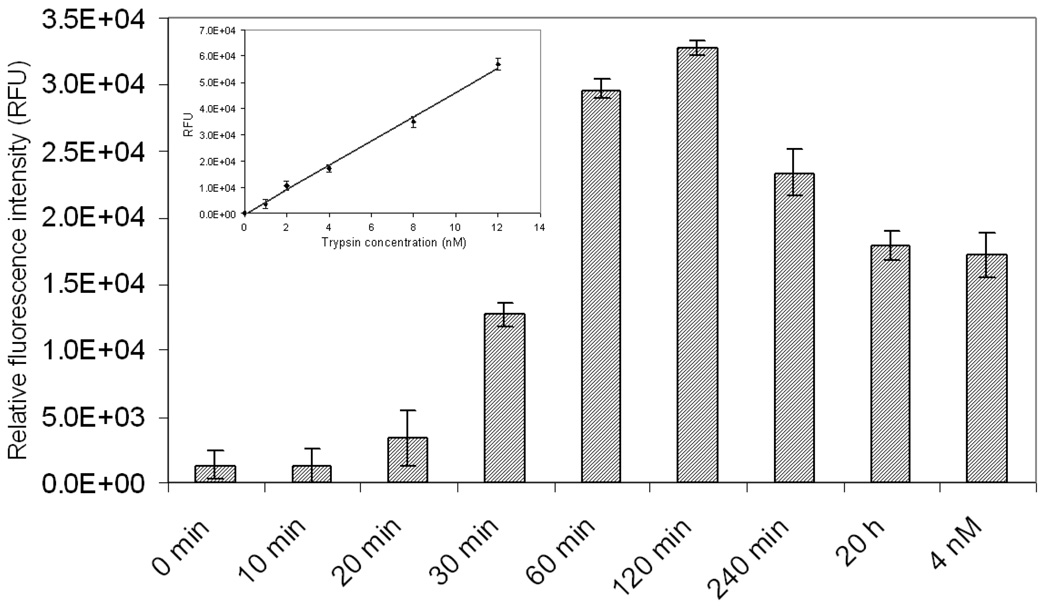
To test whether trypsinogen can be activated in pancreatic cancer cells, we first monitored trypsin activity in cell lysates of MIA PaCa-2 cells following induction with 10 nM caerulein at different time points using commercially available trypsin peptide substrate, Boc-Gln-Ala-Arg-AMC. The relative fluorescence signal change of this substrate at 450 nm can be observed as a function of activation time (Figure 5). Trypsinogen was activated and achieved the highest level following caerulein stimulation for 60 to 120 min based on the relative fluorescence unit change of trypsin peptide substrate. Using 4 nM trypsin as a positive reference, trypsinogen was activated to reach a trypsin level of 6 nM following caerulein induction for 1 h.
Figure 5.
Trypsin activity determination in cell lysates using trypsin peptide substrate (Boc-Gln-Ala- Arg-AMC). The cultured MIA PaCa-2 cells following 10 nM caerulein at various time intervals (0, 10, 20, 30, 60, 120, 240 min and 20 h) were subjected to cell lysis to extract trypsin. Trypsin activity was detected through incubation with trypsin substrate kit, Boc-Gln-Ala-Arg-AMC, at various time points following induction, which revealed that trypsin was activated and reached to the highest level following caerulein induction for 1 or 2 h based on relative fluorescence unit change of trypsin substrate. Using 4 nM trypsin as a positive reference, trypsinogen was activated to reach a rypsin level of 6 nM following caerulein induction for 1 h. The change in relative fluorescence units is also observed to be a linear correlation with active trypsin concentrations.
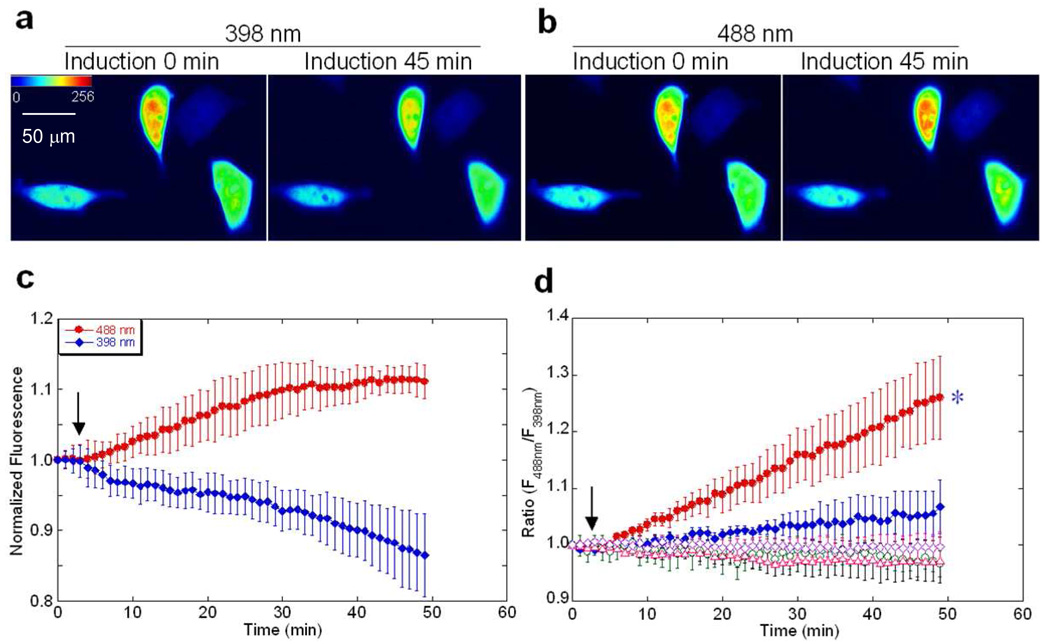
We then applied our developed sensor to track trypsin activity in living pancreatic cancer cells. The EGFP-T1 sensor was transfected into MIA PaCa-2 cells using an established method with lipofectamine (31). The cells transfected with EGFP-T1 and the controls (EGFP-wt, EGFP-T1m or EGFP-T1 incubated with leupeptin) exhibited strong fluorescence in the cytosol following 16 to 48 hours of expression. Upon zymogen activation by 10 nM caerulein, the fluorescence emission signal of EGFP-T1 at 510 nm gradually decreased at 398 nm excitation while concurrently increasing at 488 nm excitation (Figure 6a, 6b and 6c), resulting in a ratiometric emission change greater than 25% (Figure 6d). In contrast, the fluorescence emission ratio of EGFP-wt, EGFP-T1m or EGFP-T1 with SBTI inhibition remained unchanged. The ratiometric fluorescence change of EGFP-T1 sensor is significantly different from that of EGFP-wt (*P <0.05). Trypsinogen activation using 10 nM caerulein-induced in living cells (n = 12) exhibited a half-life time of 15–20 min (~80 % cells exhibit positive response), which was consistent with the results obtained using the peptide kit to monitor trypsin activity in cell lysates. By comparing enzymatic activities of different trypsin concentrations under identical experimental conditions, trypsinogen activation in pancreatic cells at the maximum stimulation time of one hour by 10 nM caerulein was determined to be approximately 6 nM. This activation is strongly dependent on caerulein concentration (Figure 6d). Therefore, our trypsin sensor can detect enzymatic activation in living cells in real time.
Figure 6.
Fluorescence imaging of EGFP-T1 in living cells. The representative images of MIA PaCa- 2 cells transfected with EGFP-T1, emitted at 510 nm with excitation of 398 nm (a) and 488 nm (b) before and after incubation with 10 nM caerulein. Normalized fluorescence (NF) intensity change (c) at 510 nm increased with excitation at 488 nm and decreased with excitation at 398 nm upon activation by caerulein induction at different times. The arrow indicates the time point of caerulein induction. Relative fluorescence change ratio (d) at 510 nm with excitation of 488/398 nm of MIA PaCa-2 cells transfected with EGFP-T1 following induction with 10 nM caerulein (●) or 5 nM caerulein (◆) exhibited significant increase, whereas the controls, EGFP-wt (○), EGFP-T1m (△) and EGFP-T1 with SBTI inhibition (◊) exhibited no change. After induction with 10 nM caerulein for 45 min, the ratiometric change in fluorescence of EGFP-T1 was significantly different from that of the control, EGFP-wt (*P<0.05).
To further examine the location of the detected trypsin signal, we applied confocal microscope and immunofluorescence by anti-trypsin and anti-GFP antibodies to evaluate the subcellular locations of trypsin and EGFP-T1, respectively. Trypsin generation following 10 nM caerulein stimulation (Supporting Figure S4a) and EGFP-T1 expression (Supporting Figure S4b) in the cytosol of MIA PaCa-2 cells was clearly observed through their corresponding antibodies. As shown in Figure S4, the fluorescence signals of trypsin antibody overlap well with that of EGFP antibody, suggesting that EGFPT1 co-localized with trypsin in the cytosol under both permeable and non-permeable conditions. The results indicated that trypsinogen is activated in the cytosol of cells, rather than on the cell surface. This novel finding for trypsinogen activation possibly provides further explanation or evidence that cytosolic events regulate zymogen activation in a reconstituted pancreatic acinar cell system (1). Since our sensor is able to detect trypsin inhibition by leupetin in vitro (Figure 3b), further application of developed trypsin sensors to other zymogen activation model systems might provide insights into the mechanism of trypsin activation and inhibition in cells.
In summary, we have reported a novel strategy to develop protease sensors by grafting an enzymatic cleavage linker onto an EGFP scaffold at different locations hypothesized to be sensitive to optical signal changes originating within the chromophore. Our EGFP-based protease sensors exhibit a large dynamic change that allows quantitative measurement of enzymatic actions. In addition, the measured kinetic parameters of our protease sensors are comparable to or better than currently available trypsin kits. Further, our trypsin sensors demonstrate strong enzymatic specificity, especially to cathepsin B and tryptase, which are essential for examining the mechanisms of zymogen activation. Furthermore, we reported the first direct observation of trypsinogen activation in MIA PaCa-2 cancer cells by caerulein stimulation even though this cell line lacks zymogen granules or cytosolic vacuole structures (14). Our results correlate well with previous reports related to elevated mRNA levels of trypsinogen (15) and trypsin inhibitors (16). Moreover, we have shown that the location of trypsinogen activation occurs within the cancer cells, shedding light on our new insight regarding zymogen activation and inhibition. Zymogen granules, vacuoles or the combination of cytosol and cellular organelles of pancreatic acinar cells were previously reported (2, 8–10). Currently, we are applying this developed sensor to further investigate the dynamic process and mechanisms for trypsinogen activation at different cellular locations (Supporting Figure S5). We anticipate that further development and application of EGFP-based sensors with strong enzymatic specificity will facilitate our understanding of protease actions in living systems. Finally, since MIA PaCa-2 cancer cells have been widely used for understanding apoptosis, cancer metastasis and drug effects, knowledge gained from zymogen activation and inhibition is expected to have an impact on future development of cancer drugs, especially against pancreatic cancers.
Supplementary Material
The experimental procedures of expression and purification, spectral properties, dynamic range calculation for optical signal change, inhibition of cleavage by leupeptin, cleavage specificity of EGFP-based trypsin sensors, statistical analysis and the supporting figures of optical properties and inhibition effects on the cleavage of trypsin sensors are available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
We would like to thank Robert Wohlhueter, Dan Adams, Michael Kirberger and Wei Yang for their critical reviews of this manuscript and helpful discussions; Kevin Francis for helpful discussions of enzyme kinetics study; Lily Yang at Emory University for the contribution of cell lines; Shenghui Xue for cell culture and imaging and other members of Dr. Yang’s research group for their helpful discussions and suggestions.
Abbreviations
- EGFP
enhanced green fluorescent protein
- PCR
polymerase chain reaction
- EGFP-wt
wild type EGFP
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- MALDI-MS
matrix assisted laser desorption/ionization-mass spectrometry
- SBTI
soybean trypsin inhibitor
- RFU
relative fluorescence units
- FITC
fluorescein isothiocyanate
- pNA
p-nitroanilide
- AMC
7-amino-4- methylcoumarin
Footnotes
This work is supported in part by following sponsors: NIH GM 62999-1, GM-70555 to JJY; GSU Molecular Basis of Disease Predoctoral Fellowships to NC; Career Award MCB-0545712 from NSF and PRF 47636-AC4 to GG.
References
- 1.Thrower EC, Diaz de Villalvilla AP, Kolodecik TR, Gorelick FS. Zymogen activation in a reconstituted pancreatic acinar cell system. Am J Physiol Gastrointest Liver Physiol. 2006;290:G894–G902. doi: 10.1152/ajpgi.00373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raraty M, Ward J, Erdemli G, Vaillant C, Neoptolemos JP, Sutton R, Petersen OH. Calcium-dependent enzyme activation and vacuole formation in the apical granular region of pancreatic acinar cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13126–13131. doi: 10.1073/pnas.97.24.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherwood MW, Prior IA, Voronina SG, Barrow SL, Woodsmith JD, Gerasimenko OV, Petersen OH, Tepikin AV. Activation of trypsinogen in large endocytic vacuoles of pancreatic acinar cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5674–5679. doi: 10.1073/pnas.0700951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirota M, Ohmuraya M, Baba H. The role of trypsin, trypsin inhibitor, and trypsin receptor in the onset and aggravation of pancreatitis. J Gastroenterol. 2006;41:832–836. doi: 10.1007/s00535-006-1874-2. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg DM. Proteases in the evaluation of pancreatic function and pancreatic disease. Clin Chim Acta. 2000;291:201–221. doi: 10.1016/s0009-8981(99)00229-6. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar FH, Banerjee S, Li Y. Pancreatic cancer: Pathogenesis, prevention and treatment. Toxicol Appl Pharmacol. 2006 doi: 10.1016/j.taap.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruger B, Albrecht E, Lerch MM. The role of intracellular calcium signaling in premature protease activation and the onset of pancreatitis. Am J Pathol. 2000;157:43–50. doi: 10.1016/S0002-9440(10)64515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mithofer K, Fernandez-del Castillo C, Rattner D, Warshaw AL. Subcellular kinetics of early trypsinogen activation in acute rodent pancreatitis. Am J Physiol. 1998;274:G71–G79. doi: 10.1152/ajpgi.1998.274.1.G71. [DOI] [PubMed] [Google Scholar]
- 9.Hofbauer B, Saluja AK, Lerch MM, Bhagat L, Bhatia M, Lee HS, Frossard JL, Adler G, Steer ML. Intra-acinar cell activation of trypsinogen during caerulein-induced pancreatitis in rats. Am J Physiol. 1998;275:G352–G362. doi: 10.1152/ajpgi.1998.275.2.G352. [DOI] [PubMed] [Google Scholar]
- 10.Otani T, Chepilko SM, Grendell JH, Gorelick FS. Codistribution of TAP and the granule membrane protein GRAMP-92 in rat caerulein-induced pancreatitis. Am J Physiol. 1998;275:G999–G1009. doi: 10.1152/ajpgi.1998.275.5.G999. [DOI] [PubMed] [Google Scholar]
- 11.Ulrich AB, Schmied BM, Standop J, Schneider MB, Pour PM. Pancreatic cell lines: a review. Pancreas. 2002;24:111–120. doi: 10.1097/00006676-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Basu A, Castle VP, Bouziane M, Bhalla K, Haldar S. Crosstalk between extrinsic and intrinsic cell death pathways in pancreatic cancer: synergistic action of estrogen metabolite and ligands of death receptor family. Cancer Res. 2006;66:4309–4318. doi: 10.1158/0008-5472.CAN-05-2657. [DOI] [PubMed] [Google Scholar]
- 13.Rosetti M, Tesei A, Ulivi P, Fabbri F, Vannini I, Brigliadori G, Amadori D, Bolla M, Zoli W. Molecular characterization of cytotoxic and resistance mechanisms induced by NCX 4040, a novel NO-NSAID, in pancreatic cancer cell lines. Apoptosis. 2006;11:1321–1330. doi: 10.1007/s10495-006-6986-x. [DOI] [PubMed] [Google Scholar]
- 14.Madden ME, Heaton KM, Huff JK, Sarras MP., Jr Comparative analysis of a human pancreatic undifferentiated cell line (MIA PaCa-2) to acinar and ductal cells. Pancreas. 1989;4:529–537. doi: 10.1097/00006676-198910000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa M, Matsuura N, Higashiyama K, Mori T. Expression of pancreatic secretory trypsin inhibitor in various cancer cells. Res Commun Chem Pathol Pharmacol. 1987;55:137–140. [PubMed] [Google Scholar]
- 16.Swarovsky B, Steinhilber W, Scheele GA, Kern HF. Coupled induction of exocrine proteins and intracellular compartments involved in the secretory pathway in AR4-2J cells by glucocorticoids. Eur J Cell Biol. 1988;47:101–111. [PubMed] [Google Scholar]
- 17.Zou J, Hofer AM, Lurtz MM, Gadda G, Ellis AL, Chen N, Huang Y, Holder A, Ye Y, Louis CF, Welshhans K, Rehder V, Yang JJ. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry. 2007;46:12275–12288. doi: 10.1021/bi7007307. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Cao Z, Yan H, Wood WC. Coexistence of high levels of apoptotic signaling and inhibitor of apoptosis proteins in human tumor cells: implication for cancer specific therapy. Cancer Res. 2003;63:6815–6824. [PubMed] [Google Scholar]
- 19.Halangk W, Sturzebecher J, Matthias R, Schulz HU, Lippert H. Trypsinogen activation in rat pancreatic acinar cells hyperstimulated by caerulein. Biochim Biophys Acta. 1997;1362:243–251. doi: 10.1016/s0925-4439(97)00082-3. [DOI] [PubMed] [Google Scholar]
- 20.Allen MD, DiPilato LM, Rahdar M, Ren YR, Chong C, Liu JO, Zhang J. Reading dynamic kinase activity in living cells for high-throughput screening. ACS Chem Biol. 2006;1:371–376. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- 21.Criddle DN, McLaughlin E, Murphy JA, Petersen OH, Sutton R. The pancreas misled: signals to pancreatitis. Pancreatology. 2007;7:436–446. doi: 10.1159/000108960. [DOI] [PubMed] [Google Scholar]
- 22.Greenbaum LM, Hirshkowitz A, Shoichet I. The activation of trypsinogen by cathepsin B. J Biol Chem. 1959;234:2885–2890. [PubMed] [Google Scholar]
- 23.Rudolf R, Mongillo M, Rizzuto R, Pozzan T. Looking forward to seeing calcium. Nat Rev Mol Cell Biol. 2003;4:579–586. doi: 10.1038/nrm1153. [DOI] [PubMed] [Google Scholar]
- 24.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 25.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felber LM, Cloutier SM, Kundig C, Kishi T, Brossard V, Jichlinski P, Leisinger HJ, Deperthes D. Evaluation of the CFP-substrate-YFP system for protease studies: advantages and limitations. Biotechniques. 2004;36:878–885. doi: 10.2144/04365PT04. [DOI] [PubMed] [Google Scholar]
- 27.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 28.Mitra RD, Silva CM, Youvan DC. Fluorescence resonance energy transfer between blue-emitting and red-shifted excitation derivatives of the green fluorescent protein. Gene. 1996;173:13–17. doi: 10.1016/0378-1119(95)00768-7. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 30.Tung CH, Mahmood U, Bredow S, Weissleder R. In vivo imaging of proteolytic enzyme activity using a novel molecular reporter. Cancer Res. 2000;60:4953–4958. [PubMed] [Google Scholar]
- 31.Zou J, Ye Y, Welshhans K, Lurtz M, Ellis AL, Louis C, Rehder V, Yang JJ. Expression and optical properties of green fluorescent protein expressed in different cellular environments. J Biotechnol. 2005;119:368–378. doi: 10.1016/j.jbiotec.2005.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The experimental procedures of expression and purification, spectral properties, dynamic range calculation for optical signal change, inhibition of cleavage by leupeptin, cleavage specificity of EGFP-based trypsin sensors, statistical analysis and the supporting figures of optical properties and inhibition effects on the cleavage of trypsin sensors are available free of charge via the Internet at http://pubs.acs.org.