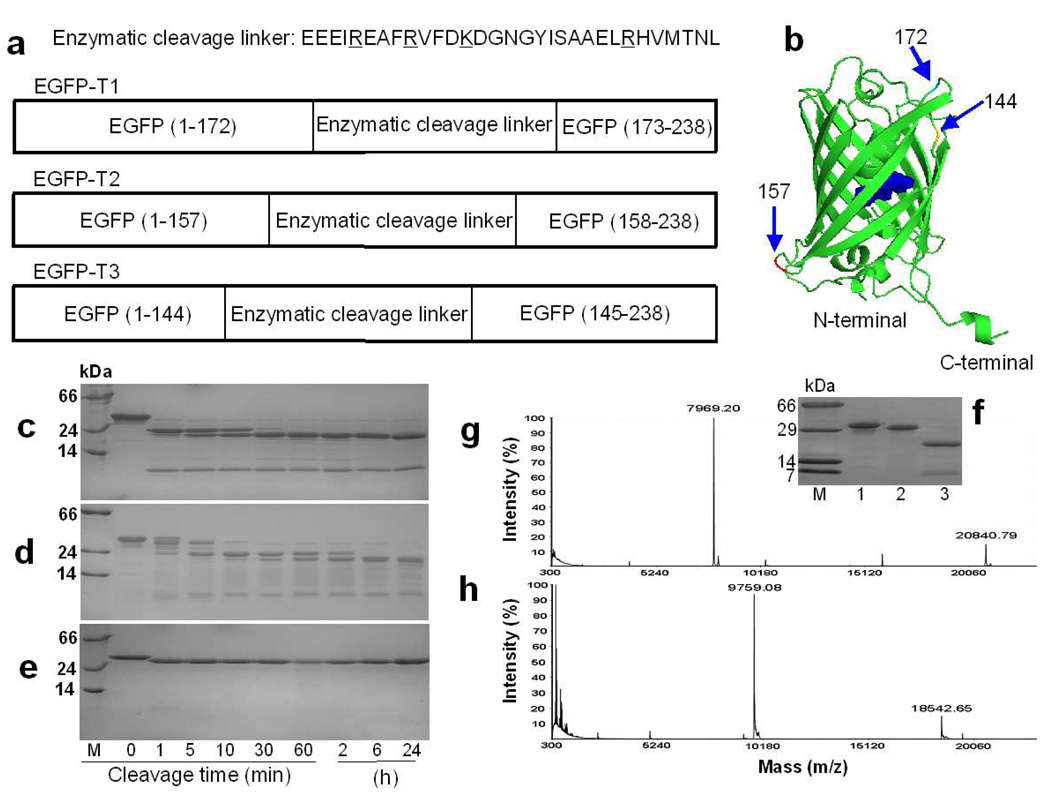
Figure 1.
The design and cleavage confirmation of trypsin sensors. a) Three EGFP-based trypsin sensors with grafted cleavable linkers at position 144, 157 and 172. Trypsin cleavage occurs following residues R or K (underlined in sequence). b) Insertion locations for trypsin linker in EGFP. SDSPAGE gels show the cleavage products of EGFP-T1 (c), EGFP-T2 (d), and EGFP-wt (e) at different digestion time intervals. M is the protein marker. Lane 1, 2, 3, 4, 5, 6, 7, 8 and 9 are samples following trypsin digestion for 0 min, 1 min, 5 min, 10 min, 30 min, 60 min, 2 h, 6 h, and 24 h at room temperature, respectively. f) To block the trypsin cleavage site, all Arg and Lys in the cleavage linker of EGFP-T1 were mutated into Ala to obtain EGFP-T1m. This mutation prevented cleavage in the inserted linker of EGFP-T1m. M is the protein marker. Lane 1, 2 and 3 are samples of EGFP-T1m without tryspin digestion, EGFP-T1m with trypsin digestion and EGFP-T1 with trypsin digestion, respectively. The product fragments of EGFP-T1 (g) and EGFP-T2 (h) cleaved by trypsin were identified by MALDI-MS.