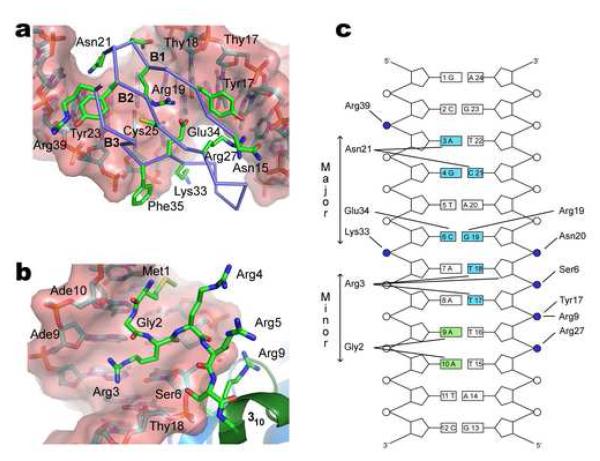
Figure 3. Mechanism of DNA binding.
(a) Expanded view of the major groove interface. Beta-sheet strands B1 (Leu16-Ile18), B2 (Tyr24-Arg27) and B3 (Glu34-Gly38) insert into the major groove. The side chains of Arg19 and Glu34 contact the Cyt6-Gua19 base pair and simultaneously form a salt-bridge. The carboxyl group of Glu34 interacts with the N4 atom of Cyt6 and the guanidine group of Arg19 donates a hydrogen-bond to the O6 atom of Gua19. The side chains of Asn20 and Lys33 contact phosphate groups of Glu19 and Ade7, respectively. The side chain of Asn21 is juxtaposed with the Gua4-Cyt21 base step in the major groove, stabilizing the binding interface. (b) Expanded view of the minor groove interface and role of the amino-terminus in DNA binding. The side chain of Met1, the backbone residues Met1-Gly2 and the side chain of Arg3 are deeply inserted into the minor groove. Gly2 contacts the Ade9-Ade10 base-step and side chain of Arg3 contacts the Thy17-Thy18 base step. Arginine residues 3, 4, 5, 9 and 10 and the N-terminal amino group are favorably positioned for electrostatic interactions with the phosphodiester backbone adjacent to the minor groove interface and the 310 helix. (c) Schematic summarizing the protein-DNA contacts in the structure of the IntN-DNA complex. Phosphodiester linkages are shown as circles, those that are contacting by IntN highlighted in blue. Bases shaded blue and green are contacted by the protein from the major- and minor-groove, respectively. A hydrogen bond is considered to be present when potential donor and acceptor atoms are separated by less than 3 Å. Salt-bridge interactions occur when appropriately charged groups are separated by less than 4.5 Å. Interactions shown in the figure occur in >40% of the structures within the ensemble.