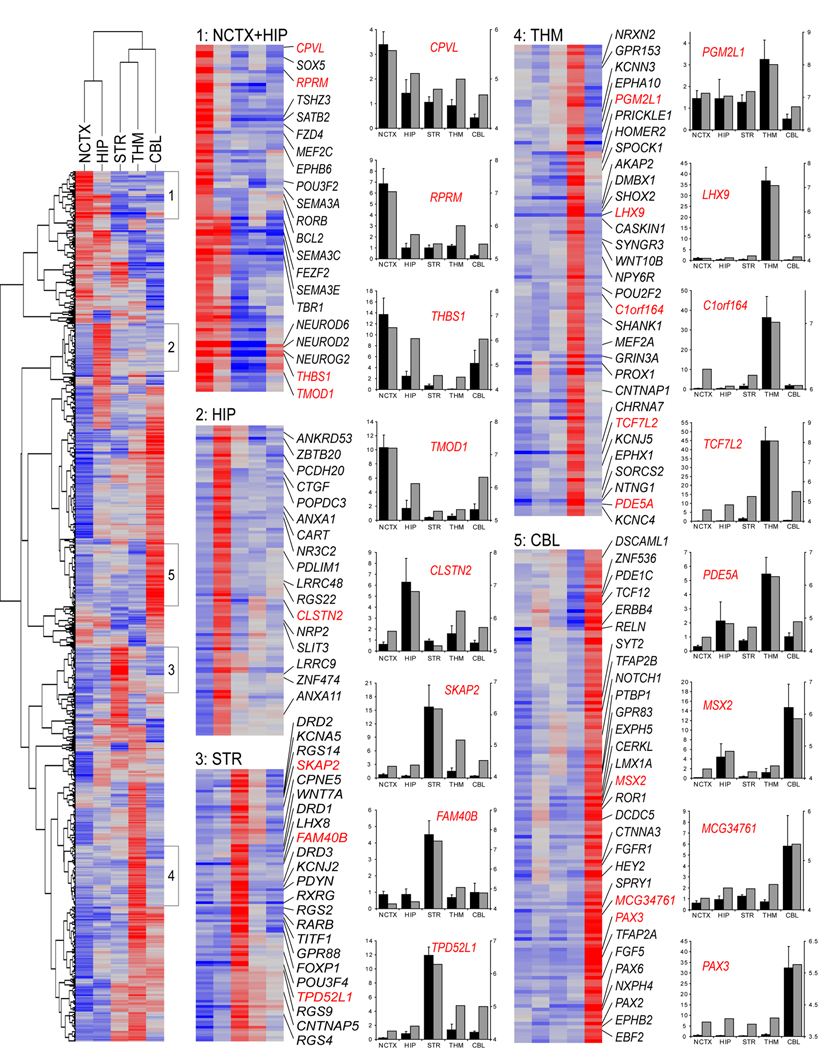
Figure 2. Unsupervised Hierarchical Clustering and qRT-PCR Validation of Selected Genes Differentially Expressed Between Brain Regions.
Genes with FDR<10−5 and greater than twofold maximum expression difference were clustered using uncentered correlation and average linkage. Selected highly correlated (r>0.75) clusters of genes with the most restricted expression patterns, indicated in the heatmap at left, are shown in detail at right, with representative validated genes labeled in red. Red is higher expression, blue is lower expression. Complete gene lists for these clusters are given in Table S6. Bar graphs show qRT-PCR (black bars) next to Exon Array data (grey bars) for representative validated genes. Bar graphs represent fold-change relative to the average of all samples for qRT-PCR (black bars; mean±SEM) and median normalized log expression level for Exon Array (grey bars).