Fig. 3.
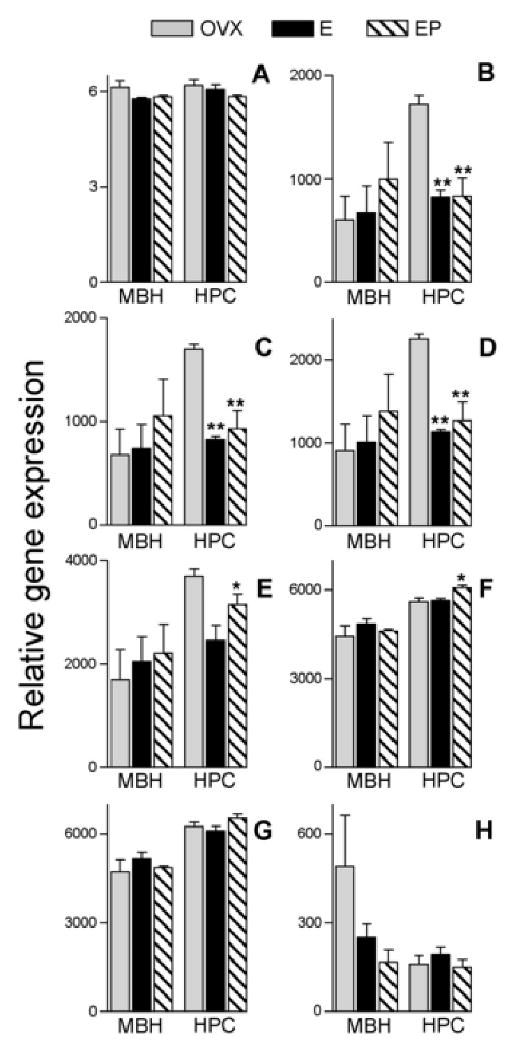
Mean and SEM of Antilogarithms to RMA-normalized rhesus macaque GeneChip® data for probesets currently featured under NetAffx™ query “ACTB”. Data are from ovariectomized females receiving hormone replacement. Probesets are ordered from 5′ to 3′ with respect to alignment on the M. mulatta β-actin mRNA sequence listed under RefSeq transcript ID NM_001033084 (see Figure 2); A = MmunewRS.18.1.S1_at; B = AFFX-Mmu-actin-5_at; C = AFFX-Mmu-actin-M_at; D = AFFX-Mmu-actin-M_x_at; E = MmunewRS.624.1.S1_s_at; F = AFFX-Mmu-actin-3_s_at; G = MmugDNA.28776.1.S1_s_at; H = MmurRNA.1.1.S1_at. MBH = arcuate nucleus of the medial basal hypothalamus, HPC = hippocampus, OVX = ovariectomized controls, E = Estrogen replacement, EP = Estrogen + Progesterone replacement. Asterisks represent significant differences in expression relative to the OVX group and show results of Newman-Keuls post hoc test following one-way ANOVA conducted on log2 transformed RMA-normalized values within each tissue. *p< 0.05, **p< 0.01.
