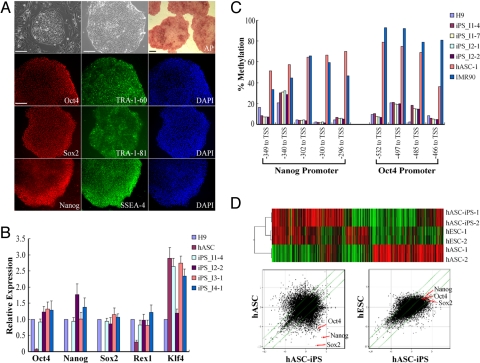
Fig. 2.
Characterization of hASC-iPS cells. (A) Immunostaining of hASC-iPS cell colonies with common hES cell markers. The two phase contrast microscopies show a typical hASC-iPS cell colony growing on MEF feeder cells and feeder-free Matrigel surface, respectively. (Scale bars, 100 μm.) (B) Quantitative-PCR analyzing pluripotency gene expression level within hASCs and hASC-iPS cells relative to those in H9 hES cells. iPS_I1–4 denotes iPS cell line #4 derived from individual 1. (C) Bisulphite pyrosequencing measuring methylation status within the promoter region of Oct4 and Nanog genes in H9 hES cells, hASC-iPS cells, hASCs, and IMR90 cells. TSS, transcription start site. (D) Microarray data comparing global gene expression profiles of hASCs, hASC-iPS cells, and hES cells. Upper panel, heat map and hierarchical clustering analysis by Pearson correlation showing hASC-iPS cells are similar to hES cells and distinct from hASCs. Lower panel, scatter plots comparing global gene expression patterns between hASCs, hASC-iPS cells, and hES cells. Highlighted are the pluripotency genes Oct4, Sox2, and Nanog (red arrows). The green diagonal lines indicated linear equivalent and 5-fold changes in gene expression levels between paired samples.