Abstract
Members of the Acr3 family of arsenite permeases confer resistance to trivalent arsenic by extrusion from cells, with members in every phylogenetic domain. In this study bacterial Acr3 homologues from Alkaliphilus metalliredigens and Corynebacterium glutamicum were cloned and expressed in Esch e richia coli. Modification of a single cysteine residue that is conserved in all analyzed Acr3 homologues resulted in loss of transport activity, indicating that it plays a role in Acr3 function. The results of treatment with thiol reagents suggested that the conserved cysteine is located in a hydrophobic region of the permease. A scanning cysteine accessibility method was used to show that Acr3 has 10 transmembrane segments, and the conserved cysteine would be predicted to be in the fourth transmembrane segment.
Arsenic is a carcinogen that ranks first on the Superfund List of Hazardous Substances (www.atsdr.cdc.gov). As a consequence of its environmental ubiquity, nearly every organism, from bacteria to humans, has genes that confer resistance to arsenic (1). The most common mechanism of arsenite resistance is efflux from cells catalyzed by members of three unrelated families of transporters. Homologues of the Mrp members of the ATP-binding cassette superfamily catalyze ATP-dependent pumping of As(III)-thiol complexes out of the cytosol. These include Mrp1 and Mrp2 in mammals that extrude As(GS)3 into blood or bile (2), Ycf1p in yeast that extrudes As(GS)3 into the vacuole (3), and PgpA in Leishmania that extrudes the As(III)-trypanothione complex into intracellular compartments (4). These pumps are generalized resistance pumps and are not specific for arsenite. In contrast, ArsB, the first identified member of the second family of arsenite efflux proteins, has the physiological role of conferring resistance to inorganic As(III) and Sb(III) (5, 6). The best characterized member of the ArsB family is that encoded by the arsRDABC operon of the conjugative R-factor R773 of Escherichia coli. ArsB is widespread in bacteria and archaea. It has 12 membrane-spanning segments (7), which is similar to members of the Major Facilitator Superfamily (8). It transports As(III) but has higher affinity for Sb(III). ArsB is an antiporter that catalyzes the exchange of trivalent metalloid for protons, coupling arsenite efflux to the electrochemical proton gradient (9).
The third arsenic resistance transporter is Acr3, which is a member of the BART (bile/arsenite/riboflavin transporter) superfamily and includes members found in bacteria, archaea, and fungi and is more widely distributed than members of the ArsB family (10) (supplemental Fig. 1). Homologues have recently been identified in plant (Pteris vittata, NCBI accession number ACN65413) and animal genomes (Danio rerio, NCBI accession number XP_001921075). Unfortunately, the literature is confused by the fact that many members of the Acr3 family are annotated as ArsB even though they exhibit no significant sequence similarity to ArsB. The first identified member of this family is encoded by the ars operon of the skin (sigK intervening) element in the chromosome of Bacillus subtilis (11). The membrane topology of the B. subtilis Acr3 was recently investigated using translational fusions, but the results could not distinguish between 8 and 10 transmembrane-spanning segments (TMs)2 (12). Fungal members of this family include the Saccharomyces cerevisiae Acr3p metalloid efflux protein (3, 13). Interestingly, yeast Acr3p appears to be selective for As(III) over Sb(III), which is surprising considering the similarity in chemical properties between the two metalloids. The properties of a more distant homologue from Shewanella oneidensis was examined recently (14). The S. oneidensis homologue confers resistance to arsenate but not arsenite. Similarly, the purified protein binds arsenate, not arsenite, indicating that this protein is not an Acr3 orthologue.
Here we examined the properties of Acr3 orthologues from Alkaliphilus metalliredigens and Corynebacterium glutamicum (supplemental Fig. 1). A. metalliredigens is a borate-tolerant Gram-positive alkaliphile and strict anaerobe that uses reduction of metals as electron acceptors (15). It is a novel metal-reducing bacterium that is distantly related to other commonly studied iron-reducing microorganisms. The genome of A. metalliredigens QYMF (NCBI accession number NC_009633) contains two novel ars operons, arsR1Bacr3–1D1A1–1A1–2 and arsR2CBacr3–2D2A2–1A2–2. The two genes for the AmAcr3s were designated arsacr3 because they are both in ars operons and are controlled by ArsR repressors, even though they are not homologues of ArsB. Interestingly, both ars operons have genes for ArsD and two genes corresponding to the two homologous halves of ArsA, which we designate AmArsA1 and AmArsA2. ArsD is an arsenic chaperone that transfers As(III) to ArsA (16), which then interacts with ArsB to extrude As(III) from the cells in an ATP-dependent manner (6, 17, 18). Whether or how Acr3 can replace ArsB in this process is a question of considerable interest.
C. glutamicum is a Gram-positive soil bacterium that is used for commercial production of glutamate, lysine, and other amino acids, nucleotides, and vitamins and from which the genome sequence has been described (NCBI accession number NC_006958). It is highly arsenic-resistant and has three genes encoding Acr3 homologues (19). Two of the homologues are in ars operons regulated by ArsRs (arsR1Bacr3–1C1C1′ and arsR2Bacr3–2arsC2) and a third orphan gene (arsBacr3–3) that is not in an operon and may not be expressed to the same extent as the other two. (Again, the genes were misnamed arsB even though they encode Acr3 homologues.)
The genes for AmAcr3 and CgAcr3 from the ars1 operons of the respective species were cloned and expressed in the arsenite-hypersensitive E. coli strain AW3110, in which the chromosomal arsRBC operon had been deleted (20). Both conferred resistance to arsenite but not arsenate or antimonite. Examination of the sequence of Acr3 homologues from many species indicates that there is conserved cysteine residues, Cys138 in AmAcr3 and Cys129 in CgAcr3 (supplemental Fig. 1). Those and other nonconserved cysteine residues were changed by mutagenesis, and substitution of only Cys138 in AmAcr3 and Cys129 in CgAcr3 led to loss of function, suggesting that the conserved cysteine residue participates in As(III) transport. A scanning cysteine accessibility method (SCAM) (21) was used to determine the transmembrane topology of AmAcr3. SCAM analysis is preferable to the use of gene fusions because there are minimal structural changes in the membrane protein, and the sidedness of inserted cysteines can be unambiguously determined with maleimide reagents of differing membrane permeability. A series of single cysteine mutants of AmAcr3 was constructed and the reactivity of each cysteine residue assayed. The results unambiguously demonstrate that Acr3 has 10 TMs.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, Media, and Reagents
Strains and plasmids used are given in supplemental Tables 1 and 2. E. coli cells were grown in Luria-Bertani (LB) medium (22) at 37 °C supplemented with 100 μg/ml ampicillin, as required. Bacterial growth was monitored by measuring the A600 nm. The thiol reagents iodoacetamide, p-chloromercuribenzoate, 5,5′-dithiobis-(2-nitrobenzoic acid), and methyl methanethiosulfonate were from Sigma. N-Ethylmaleimide was from Pierce. Biotin-PE-maleimide (N′-[2-N-maleimido)ethyl]-N-piperazinyl-d-biotinamide hydrochloride (BM), and 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) were from Molecular Probes (Eugene, OR). Other reagents were obtained from commercial sources.
Cloning of acr3 Genes, Plasmid Construction, and Mutagenesis
The acr3 genes were cloned by PCR using Pfu DNA polymerase according to the manufacturer's directions (Stratagene, La Jolla, CA). The oligonucleotide primers for PCR are given in supplemental Table 3. For expression of acr3 from A. metalliredigens in E. coli, plasmid pTrcHis2A-AmAcr3-His, in which the Amacr3 gene is under the control of the trc promoter and has the sequence for a C-terminal His tag, was constructed. Chromosomal DNA from A. metalliredigens QYMF was provided by Matthew W. Fields (Miami University, Oxford, OH) (15). After the PCR, the 1.1-kb DNA fragment was gel-purified (Qiagen) and digested with NcoI and HindIII overnight. After gel purification, the DNA fragment was ligated into vector plasmid pTrcHis2A accordingly digested, generating plasmid pTrcHis2A-AmAcr3-His. A similar strategy was used to clone the Cgacr3 gene using chromosomal DNA from C. glutamicum strain ATCC 13032 (19) and the corresponding primers indicated in supplemental Table 3. The PCR-amplified band (1,113 bp) was subcloned into pGEM®-T Easy (Promega, Madison, WI) and sequenced. The Cgacr3 gene was then PCR-amplified using the primer pair Cgacr3-NcoI forward/HindIII reverse, and the isolated 1.1-kb band was NcoI and HindIII digested and further ligated into pTrcHis2A generating plasmid pTrcHis2A-CgAcr3-His.
Mutants in acr3 genes from both organisms were generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the primers listed in supplemental Table 4. The identity of the mutated nucleotides was confirmed by DNA sequencing. Cys129 and Cys141 in CgAcr3 were changed to C129S, C141S, C129A, and C129S/C141S. Residues Cys27, Cys91, and Cys138 in AmAcr3 were changed to C27S, C91S, C138S, C138A, C27S/C91S, and C27S/C91S/C138S. For the topological studies by cysteine-scanning mutagenesis, the AmAcr3 C27S/C91S double mutant was used as the “Cys-less” backbone, and 33 single cysteine derivatives from the Cys-less were created by PCR mutagenesis (23) (supplemental Table 2).
Resistance Assays
For arsenite resistance assays, cultures of E. coli strain AW3110 harboring plasmids were grown in LB medium overnight and diluted 100-fold into fresh LB medium containing the indicated concentrations of sodium arsenite or potassium antimonyl tartrate at 37 °C with shaking. After 6 h of growth at 37 °C, A600 nm was measured.
Topological Analysis of Acr3
SCAM analysis of the topology of AmAcr3 was performed by labeling site-directed cysteines with sulfhydryl reagents, as described (21) with a few minor modifications. Mutant Amacr3 genes were transformed into cells of E. coli strain AW3110 Δars, which lacks arsB, and grown overnight in LB medium at 37 °C, diluted 50-fold into fresh LB, and induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h, when the A600 nm reached 0.5–0.6. After induction, the cells were washed with a buffer consisting of 50 mm Tris-HCl, pH 7.5, containing 0.15 m KCl (buffer A). To block cysteines exposed in the periplasm, the membrane-impermeable thiol reagent AMS was added to the cell suspension to a final concentration of 0.2 mm for 10 min at 30 °C. Next 2 μl of 25 mm BM was added to label cysteine residues exposed in the cytosol. BM is membrane-permeable and can react with thiol groups on either side of the membrane, but cysteine residues buried in hydrophobic TMs are not labeled. The reaction was stopped after 10 min by adding 1 ml of the same buffer containing 5 mm dithiothreitol. The cells were washed once with buffer A and suspended in 2 ml of a buffer consisting of 50 mm Tris-HCl, pH 7.5, containing 20% glycerol (v/v), 0.3 m KCl, 1 mm MgCl2, and 5 mm dithiothreitol (buffer B). The cells were lysed by a single pass through a French press cell at 20,000 p.s.i., and 2.5 μl of di-isopropyl fluorophosphates/g of wet cell was added immediately. After centrifugation at 1,500 × g for 5 min to remove cell debris, the supernatant was centrifuged at 150,000 × g for 1 h, and the membrane fraction obtained was suspended in 1 ml of buffer B containing 1% Triton X-100 to solubilize the inner membrane. The mixture was incubated for 1 h at 4 °C, following which the insoluble fraction was removed by centrifugation at 150,000 × g for 1 h. The supernatant solution was mixed with 15 μl of streptavidin-agarose beads (Invitrogen) and incubated with gentle agitation for 1 h at 4 °C. The agarose beads were washed five times with 0.5 ml of buffer B and then suspended in 15 μl of SDS-PAGE sample buffer (24) and boiled for 10 min. SDS-PAGE was performed on 12% polyacrylamide gels. AmAcr3 was detected by immunoblotting using mouse anti-His tag antibody (Novagen, Madison, WI) and an enhanced chemiluminescence kit (PerkinElmer Life Sciences).
Metalloid Uptake Assays in Intact Cells
For uptake assays, 50-ml cultures were grown to A600 nm = 1 at 37 °C with aeration in LB medium. The cells were harvested, washed, and suspended in a buffer consisting of 75 mm HEPES-KOH, pH 7.5, 0.15 m KCl, and 1 mm MgSO4 (buffer C) at a density of A600 nm = 10. To initiate the transport reaction, either sodium arsenite (100 μm, final concentration) or potassium antimonyl tartrate (50 μm, final concentration) was added to 1 ml of cell suspension. Portions (0.1 ml) were withdrawn at the indicated time, filtered through 0.2-μm pore size nitrocellulose filters (Whatman), washed twice with 4 ml of buffer C, and air dried. For assays in which the cells were treated with thiol-modifying reagents, 50 ml of freshly grown cells were centrifuged and washed twice at A600 nm = 1 with buffer C lacking MgSO4 and suspended in 25 ml of the same buffer. The cells were then equilibrated at 37 °C, and an equal volume of the same buffer containing 2 mm EDTA was added. After 5 min, 10 mm MgSO4 was added. The EDTA-treated cells were harvested by centrifugation and suspended in the buffer C at a density of A600 nm = 10. Thiol-modifying reagents were added as indicated for 10 min before initiating the transport reaction by addition of 100 μm sodium arsenite. The filters were digested with 0.3 ml of concentrated HNO3 (69–70%) (EM Science, Gibbstown, NJ) at 70 °C for 30 min, allowed to cool to room temperature, and diluted with 5.7 ml of high pressure liquid chromatography grade water (Sigma) to produce a final concentration of HNO3 of ∼4%. Arsenic and antimony were quantified by inductively coupled plasma mass spectroscopy. Standard solutions were made in the range of 0.5–150 ppb in 4% HNO3 using arsenic and antimony standards (Ultra Scientific, N. Kingstown, RI).
RESULTS
Cloning of acr3 Genes from C. glutamicum and A. metalliredigens
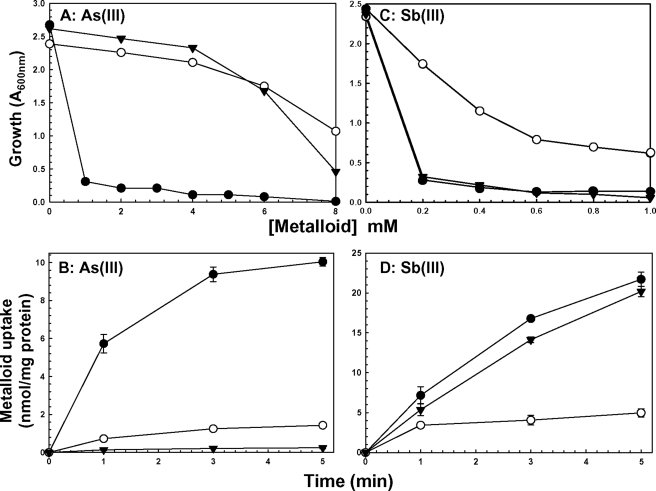
Both C. glutamicum and A. metalliredigens have two operons containing acr3 genes, and all four Acr3s exhibit considerable similarity with other Acr3 homologues (supplemental Fig. 1). Of interest is that one cysteine residue, Cys138 in AmAcr3 and Cys129 in CgAcr3, is conserved in all homologues examined. One acr3 gene was cloned from each organism and expressed in E. coli strain AW3110. Both CgAcr3 (Fig. 1, A and B) and AmAcr3 (data not shown) were functional in arsenite resistance and able to reduce the intracellular concentration of As(III), which reflects the ability to catalyze extrusion. Neither CgAcr3 (Fig. 1, C and D) nor AmAcr3 (data not shown) could confer resistance or extrude Sb(III). This is consistent with the observation that yeast ScAcr3p confers resistance to As(III) but not Sb(III) (3, 25). This is a striking finding considering the chemical similarities of the two trivalent metalloids. Note that ArsB extrudes both trivalent metalloids and confers resistance to both (Fig. 1), indicating that there might be a fundamental mechanistic difference between ArsB and Acr3.
FIGURE 1.
CgAcr3 transports and confers resistance to arsenite but not antimonite. Resistance and transport were assayed as described under “Experimental Procedures.” Assays were performed with E. coli AW3110 (Δars) bearing the following plasmids: ●, vector plasmid pKK223-3; ○, pKMB1 (arsB); ▼, pTrcHis2A-CgAcr3-His (Cgacr3). Resistance to the indicated concentrations of sodium arsenite (A) or potassium antimonyl tartrate (C) is shown. Accumulation of As(III) (B) or Sb(III) (D) was assayed. B and D, error bars represent the S.D. of three assays.
Conserved Cysteine Plays a Role in Acr3 Function
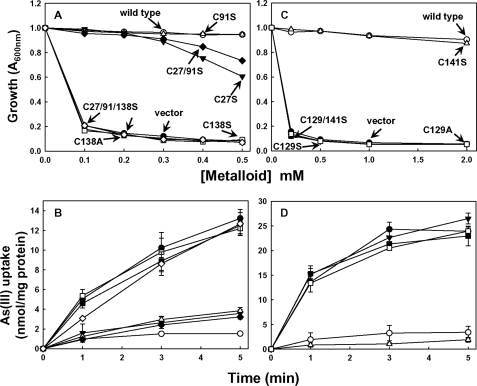
There are three cysteines in AmAcr3 (Cys27, Cys91, and Cys138). Each was altered to serine or alanine, generating single mutants C27S, C91S, C138S, and C138A, the double mutant C27S/C91S, and the triple mutant C27S/C91S/C138S. The function of each cysteine residue was analyzed by the ability to confer As(III) resistance in the arsenite-hypersensitive E. coli strain AW3110 (Fig. 2A) and to reduce the intracellular concentration of As(III) (Fig. 2B). The data show that neither Cys27 nor Cys91 are crucial for AmAcr3 function. In contrast, cells expressing mutants C138S and C138A were sensitive to and unable to extrude arsenite, suggesting that conserved Cys138 plays a role in Acr3 function. To examine further whether the conserved cysteine is required for Acr3 function, Cys129 and Cys141 in CgAcr3 were mutated to C129S, C141S, C129A, and C129S/C141S, and the effect of the substitutions on resistance (Fig. 2C) and transport (Fig. 2D) was assayed. Consistent with the results with AmAcr3, only cells expressing the Cys129 mutants of CgAcr3 exhibited arsenite sensitivity and loss of transport activity, supporting the view that the conserved cysteine plays a role in Acr3 function.
FIGURE 2.
Arsenite transport and resistance in Acr3 cysteine mutants. The cysteine residues of native AmAcr3 (A and B) or CgAcr3 (C and D) were altered by mutagenesis, and arsenite resistance (A and C) and transport (B and D) were assayed as described under “Experimental Procedures” in E. coli AW3110 (Δars) with the following plasmids. A and B, ●, vector plasmid pTrcHis2A; ○, pTrcHis2A-AmAcr3-His (Amacr3); ▼, pAmAcr3C27S-His; Δ, pAmAcr3C91S-His; ■, pAmAcr3C138S-His; □, pAmAcr3C138A-His; ♦, pAmAcr3C27/91S-His; ♢, pAmAcr3C27/91/138S-His. C and D, ●, vector plasmid pTrcHis2A; ○, pTrcHis2A-CgAcr3-His (Cgacr3); ▼, pCgAcr3C129S-His; Δ, pCgAcr3C141S-His; ■, pCgAcr3C129/141S-His; □, pCgAcr3C129A-His. B and D, error bars represent the S.D. of three assays.
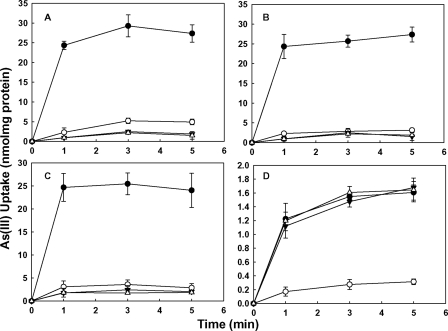
To investigate the requirement for the conserved cysteine in more detail, uptake of As(III) was assayed in cells of AW3110 expressing wild type CgAcr3 and the C141S mutant, which has only conserved Cys129 treated with several different thiol reagents (Fig. 3). The cells were first treated with EDTA to permeabilize the outer membrane and then with 5,5′-dithiobis-(2-nitrobenzoic acid) (Fig. 3B) or p-chloromercuribenzenesulfonate (Fig. 3C), which are membrane-impermeant, or the membrane-permeable thiol reagent N-ethylmaleimide (Fig. 3D) (26–28). Arsenite accumulation in cells expressing wild type CgAcr3 and the C141S mutant was not affected by 5,5′-dithiobis-(2-nitrobenzoic acid) or p-chloromercuribenzenesulfonate. In contrast, the permeable reagent N-ethylmaleimide inactivated As(III) efflux. Similarly, accumulation was inhibited by the permeant thiol-modifying reagents methyl methanethiosulfonate and iodoacetamide (data not shown). As a control, arsenite accumulation in cells of E. coli AW3110 expressing ArsB, which has no critical thiols (29), was unaffected by any of the thiol reagents. These results suggest first that the conserved cysteine in Acr3 participates in As(III) translocation and second that it may be located in a hydrophobic transmembrane region, a prediction verified by the topological analysis below.
FIGURE 3.
Effect of thiol-modifying reagents on arsenite uptake by CgAcr3. Arsenite accumulation was assayed as described under “Experimental Procedures” in EDTA-treated cells of AW3110 expressing the following: ●, vector plasmid pTrcHis2A; ○, pKMB1 (arsB); ▼, pTrcHis2A-CgAcr3-His (Cgacr3); and Δ, pCgAcr3C141S-His with the following additions: A, no addition; B, 1 mm 5,5′-dithiobis-(2-nitrobenzoic acid); C, 0.2 mm p-chloromercuribenzenesulfonate; and D, 1 mm N-ethylmaleimide. The error bars represent the S.D. of three assays.
Membrane Topology of Acr3
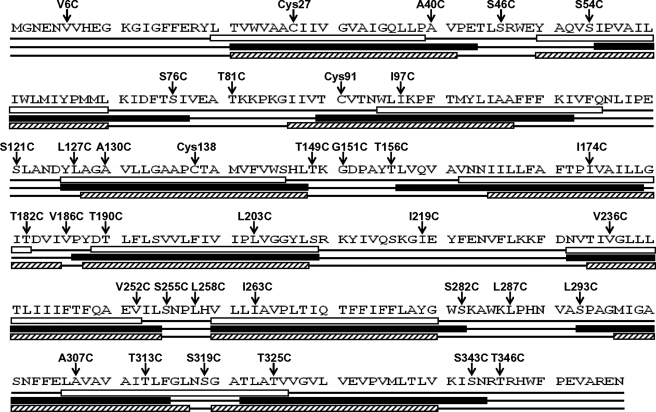
Although ArsB has 12 TM segments (7), Acr3 has been predicted to have only 10 TMs (12). We compared predictions for AmAcr3 from three different topological analysis programs, TopPred2, SOSUI, and S-TMHMM. These generated two different theoretical models, one with nine TMs (TopPred 2) and the other with 10 TMs (SOSUI and S-TMHMM) (Fig. 4). It is worth mentioning that in all predicted models, the conserved residue Cys138 is located in a transmembrane region. An incomplete topological analysis of the B. subtilis Acr3 generated using translational fusions could not distinguish between 8 and 10 transmembrane segments (12). In this study, SCAM (21) was used to determine experimentally the transmembrane topology of AmAcr3.
FIGURE 4.
Predictions of the transmembrane topology of AmAcr3. Secondary structure predictions for AmAcr3 were generated from the primary amino acid sequence (1st line) with TopPred2 (2nd line with TMs in open boxes), SOSUI (3rd line with TMs in filled boxes), and S-TMHMM (4th line with TMs in hatched boxes). Above each are cysteine residues, either native or introduced, with the conserved residue Cys138 located in TM4 with all predictions. TopPred2 predicted 9 TMs, whereas SOSUI and TMHMM predicted 10 TMs.
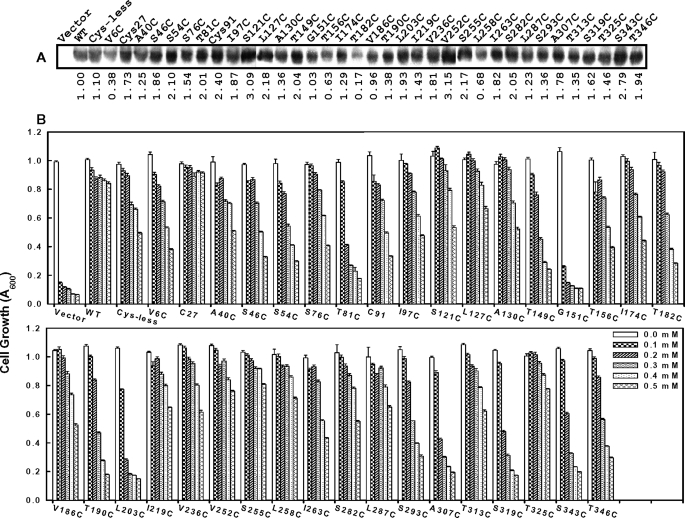
Starting with the C27S/C91S double mutant of AmAcr3, which has only Cys138, 33 single mutants containing only a single additional cysteine residue were constructed covering all predicted hydrophobic and hydrophilic regions. Cys138 is necessary for function, but as shown below, it does not react with any of the SCAM reagents, and therefore the C27S/C91S mutant in effect provides the Cys-less backbone required for SCAM analysis. The expression of each in E. coli AW3110 was examined by immunoblotting (Fig. 5A). Each of the mutants was expressed, although some (V6C, T156C, T182C, and L258C) were expressed at lower levels than wild type. Their ability to confer resistance to arsenite was determined (Fig. 5B). Except for the G151C mutant, each conferred resistance. Some mutants such as T81C, T190C, L203C, A307C, S319C, and S343C were somewhat less resistant at higher concentrations of arsenite. However, more important than the absolute levels of expression or resistance, the fact that they clearly retain function indicates that they should have an overall topology similar to that of the wild type, which makes them useful for SCAM analysis.
FIGURE 5.
Protein expression and arsenite resistance in cysteine mutants of AmAcr3. Cells of E. coli AW3110 expressing the indicated cysteine mutants of Amacr3 were assayed for AmAcr3 protein expression by immunoblotting with His tag antibody (A) and resistance to the indicated concentrations of sodium arsenite (B). Each mutant included the conserved residue Cys138, without which there is no activity. Cells with plasmid pTrcHis2A (Vector) or wild type (WT) Amacr3 were used as negative and positive controls, respectively. The Cys-less mutant retains Cys138 but has the C27S/C91S double mutation. The mutant named C27 retains Cys27 and Cys138 but has the C91S mutation, and the mutant named C91 retains Cys91 and Cys138 but has the C27S mutation. The error bars represent the S.D. of three assays. A, relative amounts of protein in a typical experiment (from three assays) compared with wild type AmAcr3 (WT) were estimated by densitometry.
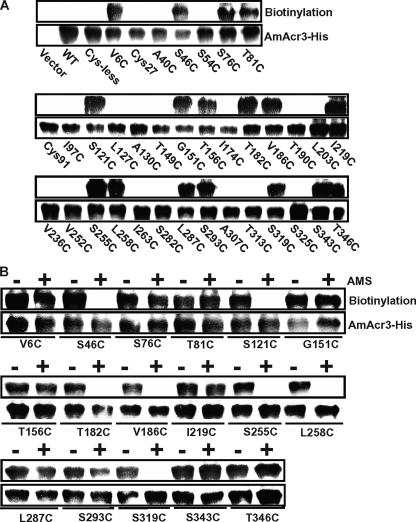
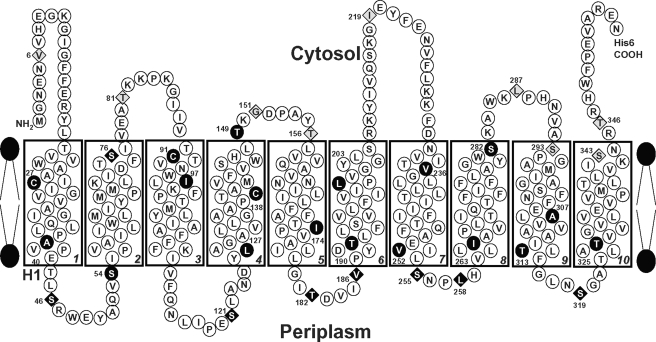
To determine topology using SCAM, the cells were first treated with BM, and the BM-labeled Acr3 proteins were pulled down with streptavidin-conjugated beads after solubilization of the membranes with Triton X-100. Biotinylated proteins were detected by immunoblotting with His tag antibody. BM is membrane-permeable and reacts with cysteine thiolates accessible on either side of the membrane but not with cysteines in TMs. The wild type, the mutants with Cys27 or Cys91, and 16 mutants with introduced cysteine residues (A40C, S54C, I97C, L127C, A130C, T149C, I174C, T190C, L203C, V236C, V252C, I263C, S282C, A307C, T313C, and T325C) did not react with BM (Fig. 6A), indicating that they are in a TM or otherwise not exposed to bulk solvent. Note that neither wild type AmAcr3 nor the Cys-less variant reacted with BM, indicating that none of the three native cysteine residues (Cys27, Cys91, and conserved Cys138) are solvent-exposed and are quite possibly in transmembrane regions. The 17 mutants that reacted with BM were analyzed further by reacting with AMS, which is impermeant and reacts with periplasmic cysteine residues. The mutants were then reacted with BM, which would react with the remaining cysteine residues that are exposed in the cytosol. AMS prevented biotinylation of seven mutants (S46C, S121C, T182C, V186C, S255C, L258C, and S319C) (Fig. 6B), indicating that these residues are exposed to the periplasm. The other 10 residues (V6C, S76C, T81C, G151C, T156C, I219C, L287C, S293C, S343C, and T346C) reacted with BM even after AMS treatment, indicating that the residues in those mutants are most likely exposed to the cytosol. These results are consistent with a topological model of 10 TMs with the N and C termini in the cytosol (Fig. 7). Note that this model makes several predictions. First, the conserved cysteine residue is located in TM4, and second that Gly151 is in a short cytosolic loop. The G151C mutant may not have sufficient flexibility to allow for proper orientation of TM4 and TM5, which may explain its inability to confer arsenite resistance.
FIGURE 6.
SCAM analysis of AmAcr3 cysteine mutants. Cysteine mutants of Amacr3 were reacted with BM (A) or BM with or without AMS pretreatment (B). The top panel in each shows a typical reaction with BM as detected by pulldown assays with streptavidin-agarose beads. The bottom panel in each shows the results of a typical assay (from three assays) of immunoblotting of membranes from cells expressing the indicated mutant Amacr3 genes. WT, wild type.
FIGURE 7.
Topology of AmAcr3. The topological model of AmAcr3 is based on the SCAM data from Fig. 4. The numbered residues were altered to cysteines. ●, BM-inaccessible residues; ♢, BM-accessible, AMS-inaccessible residues; ♦, residues accessible to both BM and AMS. The conserved Cys138 is located in TM4.
DISCUSSION
As a result of the environmental pervasiveness of arsenic, various ways of detoxification have evolved, and efflux is one of the more common mechanisms. One of the most widespread arsenic efflux proteins is Acr3, which is found in members of every kingdom. Yet little is known about the mechanism of Acr3 transport. In this study we examined the transport properties of two Acr3 permeases from two quite different bacteria, the iron-reducing alkaliphile, A. metalliredigens QYMF, and a soil bacterium, C. glutamicum. Both organisms have two ars operons, each containing an acr3 gene, and consequently, both organisms are extremely arsenic-resistant. One acr3 gene from each was cloned and expressed in a strain of E. coli in which the chromosomal arsRBC operon had been deleted and so is extremely arsenic-sensitive. Both acr3 genes complemented the arsenite sensitivity quite well, and both catalyzed arsenite efflux from cells, allowing the first in vivo analysis of Acr3 transport properties.
The first very striking observation is that Acr3 can distinguish between As(III) and Sb(III) (Fig. 1). This had been inferred from the phenotype of yeast expressing ScAcr3p but never directly demonstrated before (25). Considering metalloid chemistry, this is unexpected. Both As(III) and Sb(III) are three-coordinate metals that bind to two or three cysteines in other arsenic resistance proteins, including ArsA (30), ArsD (31), and three different ArsRs (32–34). Even ArsB, which uses no cysteines in its translocation mechanism, is not selective for As(III) over Sb(III) (29). Interestingly, a somewhat related arsenic resistance protein from a Gram-negative anaerobe, S. oneidensis, transports pentavalent arsenate and not As(III) or Sb(III) (14). Other members of the BART superfamily transport a variety of substrates, including riboflavin, bile salts, and other organic anions (10). The basis for substrate specificity in members of this superfamily is not clear. However, one feature that distinguishes the As(III) carriers from other superfamily members appears to be a conserved cysteine residue (supplemental Fig. 1). Modification of this cysteine, either by mutagenesis (Fig. 2) or chemical modification (Fig. 3), leads to loss of transport activity. A reasonable deduction is that the conserved cysteine is involved in As(III) translocation. This residue is located in the middle of a transmembrane segment (Fig. 7), and it is tempting to speculate that thiol chemistry is involved in translocation. Even though single thiols bind trivalent arsenic with low affinity, so that As(III) might bind weakly on one side of the membrane, the low affinity would facilitate release of As(III) from the carrier on the other side of the membrane.
Acr3 and ArsB are similar in that both are secondary carriers that extrude As(III) from cells. However, they differ not only in selectivity but also in transmembrane topology. ArsB has 12 TMs (7). Theoretical topological analysis of Acr3 suggested 9 or 10 TMs (Fig. 4), and experimental analysis of the B. subtilis Acr3 indicated either 8 or 10 TMs (12). However, gene fusions were used in that study, so the results give the topology of inactive truncated membrane proteins and not the native active permease. The use of SCAM provides for topological determination of intact and active membrane proteins (21). Using this method, Acr3 was shown to have 10 TMs (Fig. 7). The location of the conserved cysteine residue in TM4 is consistent with a role in As(III) translocation.
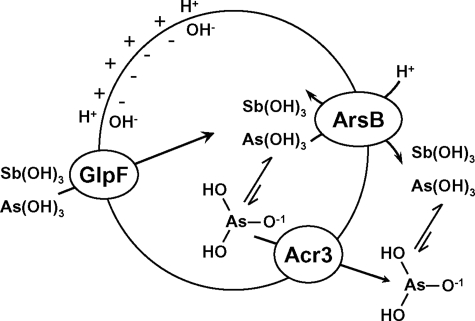
Preliminary data indicate that arsenite efflux via Acr3 is coupled to the protonmotive force,3 but how is not clear. ArsB is an antiporter in which As(OH)3 is exchanged for H+, and the counter-transport is driven by the protonmotive force. It is possible that Acr3 functions similarly. However, another possibility is that Acr3 is a uniporter that transports the arsenite anion As(OH)2O− coupled to the membrane potential, which is positive outside in cells and positive inside in membrane vesicles (Fig. 8). The pKa of As(OH)3 is 9.2, so a few percent would be ionized in the cytosol of E. coli and even more in the cytosol of alkaliphiles. In contrast, the pKa of Sb(OH)3 is 11.8; hence, at the neutral pH of the E. coli cytosol, there would essentially be no antimonite anion. This difference in pKa between the two trivalent metalloids might explain the ability of Acr3 to discriminate between As(III) and Sb(III). To elucidate the mechanism of Acr3 catalysis requires a more detailed structure-function analysis.
FIGURE 8.
Model of metalloid transport by Acr3 and ArsB. In cells of E. coli uptake of As(OH)3 or Sb(OH)3, uptake is facilitated by the GlpF channel. ArsB is an antiporter that exchanges As(OH)3 or Sb(OH)3 in exchange for a proton. Exchange of either neutral metalloid with positively charged H+ couples efflux to the protonmotive force. Acr3 is a uniporter that extrudes the As(OH)2O− anion driven by the membrane potential, positive outside. The arsenite anion is in equilibrium with the neutral acid form according to its pKa of 9.2. With a pKa of 11.8, the concentration of the antimonite anion in cytosol of E. coli is too low for significant amounts of Sb(III) to be transported by Acr3. Thus the difference in pKa of the substrates explains the difference in selectivity between Acr3 and ArsB.
Supplementary Material
Acknowledgment
We thank Matthew W. Fields, Miami University, Oxford, OH, for A. metalliredigens QYMF chromosomal DNA.
This work was supported, in whole or in part, by National Institutes of Health Grant R37 GM55425 (to B. P. R.). This work was also supported by Grants LE040A07, BIO2005-02723, and BIO2008-00519 (to L. M. M. and J. A. G.) from the Spanish Government.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1, Tables S1–S4, and additional references.
H. L. Fu and B. P. Rosen, manuscript in preparation.
- TM
- transmembrane-spanning segment
- SCAM
- scanning cysteine accessibility method
- LB
- Luria broth
- AMS
- 4-acetamido-4′-maleimidylstilbene-2-2′disulfonic acid
- BM
- biotin-PE-maleimide(N′-[2-N-maleimido)ethyl]-N-piperazinyl-d-biotinamide hydrochloride.
REFERENCES
- 1.Bhattacharjee H., Rosen B. P. (2007) in Molecular Microbiology of Heavy Metals (Nies D. H., Silver S. eds) pp. 205–219, Springer-Verlag, Inc., New York [Google Scholar]
- 2.Deeley R. G., Westlake C., Cole S. P. (2006) Physiol. Rev. 86, 849–899 [DOI] [PubMed] [Google Scholar]
- 3.Ghosh M., Shen J., Rosen B. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5001–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Légaré D., Richard D., Mukhopadhyay R., Stierhof Y. D., Rosen B. P., Haimeur A., Papadopoulou B., Ouellette M. (2001) J. Biol. Chem. 276, 26301–26307 [DOI] [PubMed] [Google Scholar]
- 5.Chen C. M., Misra T. K., Silver S., Rosen B. P. (1986) J. Biol. Chem. 261, 15030–15038 [PubMed] [Google Scholar]
- 6.Tisa L. S., Rosen B. P. (1990) J. Biol. Chem. 265, 190–194 [PubMed] [Google Scholar]
- 7.Wu J., Tisa L. S., Rosen B. P. (1992) J. Biol. Chem. 267, 12570–12576 [PubMed] [Google Scholar]
- 8.Marger M. D., Saier M. H., Jr. (1993) Trends Biochem. Sci. 18, 13–20 [DOI] [PubMed] [Google Scholar]
- 9.Meng Y. L., Liu Z., Rosen B. P. (2004) J. Biol. Chem. 279, 18334–18341 [DOI] [PubMed] [Google Scholar]
- 10.Mansour N. M., Sawhney M., Tamang D. G., Vogl C., Saier M. H., Jr. (2007) FEBS J. 274, 612–629 [DOI] [PubMed] [Google Scholar]
- 11.Sato T., Kobayashi Y. (1998) J. Bacteriol. 180, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aaltonen E. K., Silow M. (2008) Biochim. Biophys. Acta 1778, 963–973 [DOI] [PubMed] [Google Scholar]
- 13.Bobrowicz P., Wysocki R., Owsianik G., Goffeau A., Ułaszewski S. (1997) Yeast 13, 819–828 [DOI] [PubMed] [Google Scholar]
- 14.Xia X., Postis V. L., Rahman M., Wright G. S., Roach P. C., Deacon S. E., Ingram J. C., Henderson P. J., Findlay J. B., Phillips S. E., McPherson M. J., Baldwin S. A. (2008) Mol. Membr. Biol. 25, 691–705 [DOI] [PubMed] [Google Scholar]
- 15.Ye Q., Roh Y., Carroll S. L., Blair B., Zhou J., Zhang C. L., Fields M. W. (2004) Appl. Environ. Microbiol. 70, 5595–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y. F., Walmsley A. R., Rosen B. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15617–15622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey S., Dou D., Rosen B. P. (1994) J. Biol. Chem. 269, 25442–25446 [PubMed] [Google Scholar]
- 18.Dey S., Dou D., Tisa L. S., Rosen B. P. (1994) Arch. Biochem. Biophys. 311, 418–424 [DOI] [PubMed] [Google Scholar]
- 19.Ordóñez E., Letek M., Valbuena N., Gil J. A., Mateos L. M. (2005) Appl. Environ. Microbiol. 71, 6206–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlin A., Shi W., Dey S., Rosen B. P. (1995) J. Bacteriol. 177, 981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdanov M., Zhang W., Xie J., Dowhan W. (2005) Methods 36, 148–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Kirsch R. D., Joly E. (1998) Nucleic Acids Res. 26, 1848–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25.Wysocki R., Bobrowicz P., Ułaszewski S. (1997) J. Biol. Chem. 272, 30061–30066 [DOI] [PubMed] [Google Scholar]
- 26.Olsowski A., Monden I., Keller K. (1998) Biochemistry 37, 10738–10745 [DOI] [PubMed] [Google Scholar]
- 27.Zoccarato F., Cavallini L., Valente M., Alexandre A. (1999) Neurosci. Lett. 274, 107–110 [DOI] [PubMed] [Google Scholar]
- 28.Tamura N., Konishi S., Iwaki S., Kimura-Someya T., Nada S., Yamaguchi A. (2001) J. Biol. Chem. 276, 20330–20339 [DOI] [PubMed] [Google Scholar]
- 29.Chen Y., Dey S., Rosen B. P. (1996) J. Bacteriol. 178, 911–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan X., Bhattacharjee H., Rosen B. P. (2006) J. Biol. Chem. 281, 9925–9934 [DOI] [PubMed] [Google Scholar]
- 31.Lin Y. F., Yang J., Rosen B. P. (2007) J. Biol. Chem. 282, 16783–16791 [DOI] [PubMed] [Google Scholar]
- 32.Qin J., Fu H. L., Ye J., Bencze K. Z., Stemmler T. L., Rawlings D. E., Rosen B. P. (2007) J. Biol. Chem. 282, 34346–34355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi W., Dong J., Scott R. A., Ksenzenko M. Y., Rosen B. P. (1996) J. Biol. Chem. 271, 9291–9297 [DOI] [PubMed] [Google Scholar]
- 34.Ordóñez E., Thiyagarajan S., Cook J. D., Stemmler T. L., Gil J. A., Mateos L. M., Rosen B. P. (2008) J. Biol. Chem. 283, 25706–25714 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.