FIGURE 6.
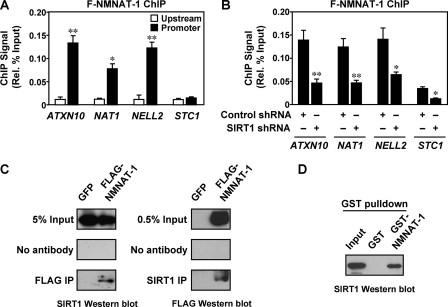
NMNAT-1 is recruited to target gene promoters by SIRT1. A, FLAG-based ChIP-qPCR analysis of NMNAT-1 localization at promoter and upstream (∼−10 kilobase) regions of target genes. MCF-7 cells expressing FLAG-NMNAT-1 were used for the FLAG ChIP assay. Error bars, S.E.; n ≥ 3 independent biological replicates. Statistical significance for differences between ChIP signals at upstream and promoter regions of the same gene was determined by a two-tailed Student's t test (*, p < 0.05; **, p < 0.01). B, FLAG-based ChIP-qPCR analysis of NMNAT-1 localization at target gene promoters in control or SIRT1 knockdown cells. Error bars, S.E.; n ≥ 3 independent biological replicates. Statistical significance for differences between ChIP signals in control and SIRT1 knockdown cells was determined by a two-tailed Student's t test (*, p < 0.05; **, p < 0.01). C, ChIP-Western analysis of interaction between SIRT1 and NMNAT-1. MCF-7 cells expressing GFP or FLAG-NMNAT-1 were used for FLAG and SIRT1 ChIP assays. The immunoprecipitated (IP) material was subjected to SIRT1 and FLAG Western blot analysis, respectively. Gels shown are representative of three independent experiments. D, GST-NMNAT-1 interaction assay with native SIRT1 from nuclear extract. SIRT1 bound to glutathione-agarose resin was detected by Western blot analysis. The gel shown is representative of three independent experiments.