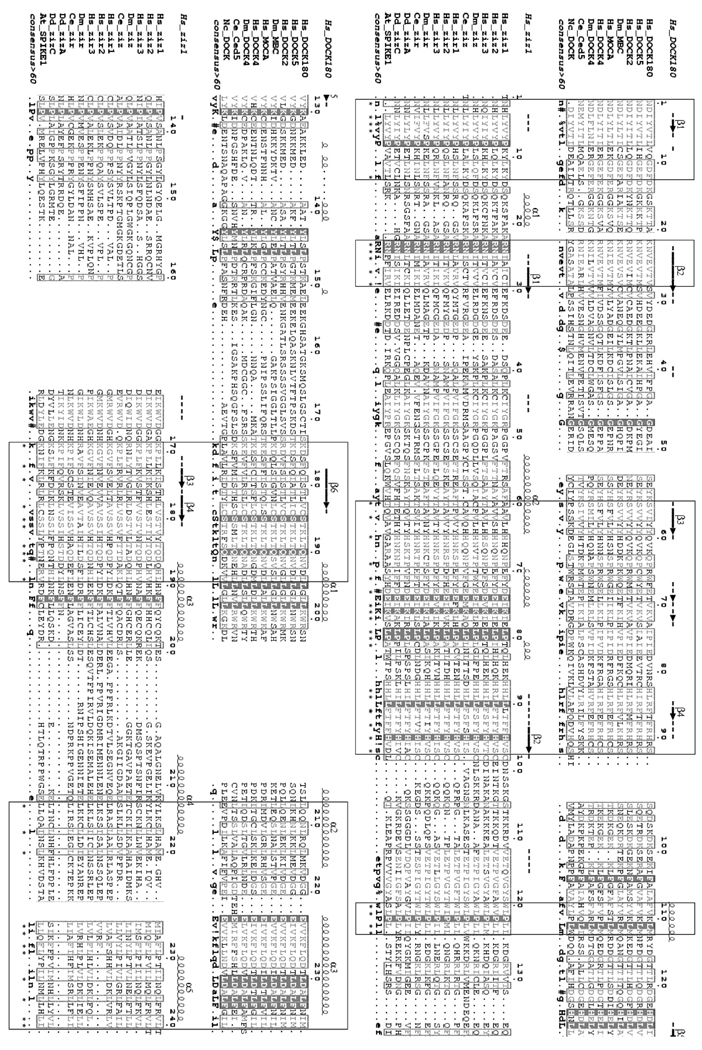
Fig. 7. Sequence alignment of CZH1 domains.
CZH1 sequences of zizimin-related and DOCK180-related proteins were aligned separately using the Multalin program and consensus sequences were generated [35]. Residues sharing >60% similarity are typed in gray while residues sharing 100% identity are presented on dark background. Secondary structure predictions for zizimin1 and DOCK180 CZH1 domains were made by the PHD program [36] and are shown on top of the alignments. The two resulting alignments were aligned together. Stretches showing resemblance to the consensus sequences are boxed and similar columns are marked by asterisks. ! represents I or V; # represents any one of N, D, Q or E; and $ represents Y or F. The numbering is according to zizimin1, or DOCK180, with 1 being residue 640 or 426, respectively. The approximate boundaries of the domain were determined as follows: The zizimin1 sequence was truncated into overlapping 200–500 amino acid segments and each segment was searched against the NCBI NR database using PSI-BLAST at an E-value threshold of 1e−2. BLAST hits were retrieved and separate multiple alignments were constructed for each segment using CLUSTALW [37]. Areas of homology between zizimin and DOCK180 proteins were identified and confined on the basis of alignment quality and secondary structure prediction. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Nc, Neurospora crassa; Dd, Dictyostelium discoideum; At, Arabidopsis thaliana.