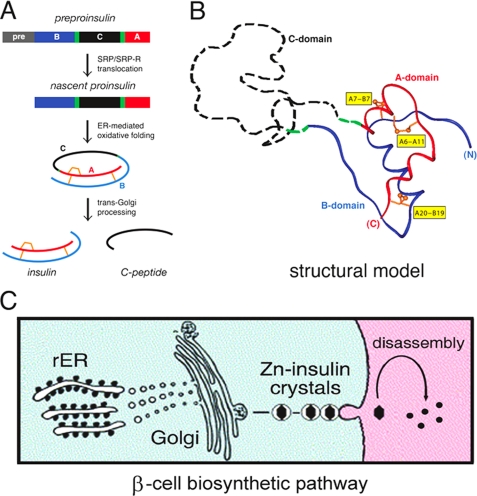
FIGURE 1.
Proinsulin and its biosynthetic pathway. A, pathway of insulin biosynthesis beginning with preproinsulin (upper): signal peptide (gray), B-domain (blue), dibasic BC junction (green), C-domain (red), dibasic CA junction (green), and A-domain (red). In the ER, the unfolded prohormone undergoes specific disulfide pairing to yield native proinsulin (center). Cleavage of BC and CA junctions by prohormone convertases (PC1 and PC2) and carboxypeptidase E leads to mature insulin and the C-peptide (lower). SRP/SRP-R, signal recognition particle/signal recognition particle receptor. B, structural model of insulin-like moiety and disordered connecting peptide (dashed black line). The A- and B-domains are shown in red and blue, respectively; the disordered connecting domain is shown by the dashed black line. Cystines are labeled in yellow boxes. C, cellular pathway of insulin biosynthesis. Nascent proinsulin folds as a monomer in the rough ER (rER; left), wherein zinc ion concentration is low; in post-Golgi granules, proinsulin is processed by cleavage of the connecting peptide to yield mature insulin, and zinc-stabilized hexamers begin to assemble. Zinc-insulin crystals are observed in secretory granules. Upon secretion into the portal circulation (right), hexamers dissociate to yield bioactive insulin monomers. Although the structure of an isolated monomer resembles that of a crystallographic protomer, marked changes in conformation may be required for receptor binding. Native hexamer assembly and induced fit of the insulin monomer may provide complementary structural adaptations to the threat of toxic protein misfolding (49, 75, 76).