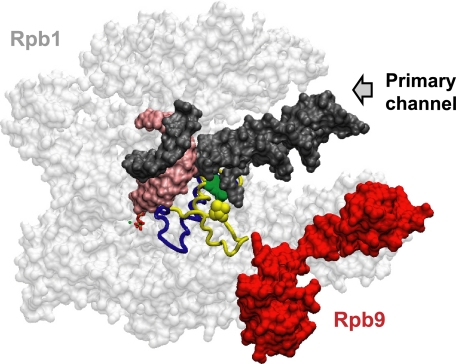
FIGURE 6.
Model describing the effect of RPB9 deletion on transcription fidelity. TEC structures of Saccharomyces cerevisiae pol II with the closed trigger loop (blue) and the open trigger loop (yellow) (PDB: 2E2H (14) and PDB: 1Y1V (22)) were aligned using the VMD software (59). Rpb1 (gray) is made semitransparent to visualize the location of the TL inside the pol II structure, and Rpb9 is shown in red. The NTP in the active center of pol II is shown on the left side of the TL. The DNA backbone and RNA are shown in dark gray and pink, respectively. The primary DNA-binding channel in pol II is indicated by the leftward arrow. The secondary channel is located at the opposite side of the structure. The Rpb9 subunit stabilizes the open conformation of the TL by interaction between the C-terminal domain of Rpb9 with the tip of the TL (yellow), similar to the proposed interaction between Rpb1-Glu1103 (green) with Rpb1-Thr1095 (yellow) (13).