Figure 3.
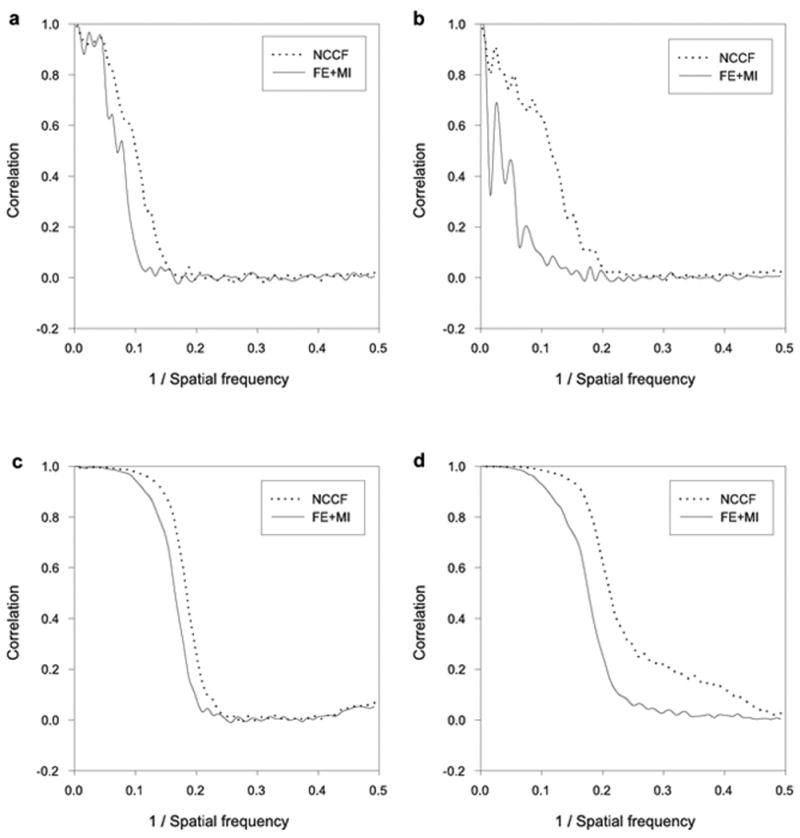
Model dependency test on synthetic data sets. Comparison of reconstructed structure resolution for NCCF and FE+MI. (a) FSC between reconstructions obtained from halves of the SF3b data when a ribosome template structure is used to align and assign Euler angles to the SF3b data. (b) FSC between reconstructions of the SF3b data and the ribosome data when a ribosome template structure is used to align and assign Euler angles to the two respective data sets. (c) FSC between reconstructions obtained from halves of the SF3b data when a SF3b template structure is used to align and assign Euler angles to the SF3b data. (d) FSC between (1) a reconstruction of the SF3b data obtained after alignment with the NCCF and FE+MI method and (2) a reconstruction of the SF3b data with no alignment error and no error in the assignment of Euler angles. A SF3b template was used to align and assign Euler angles to the SF3b data.
