Figure 7.
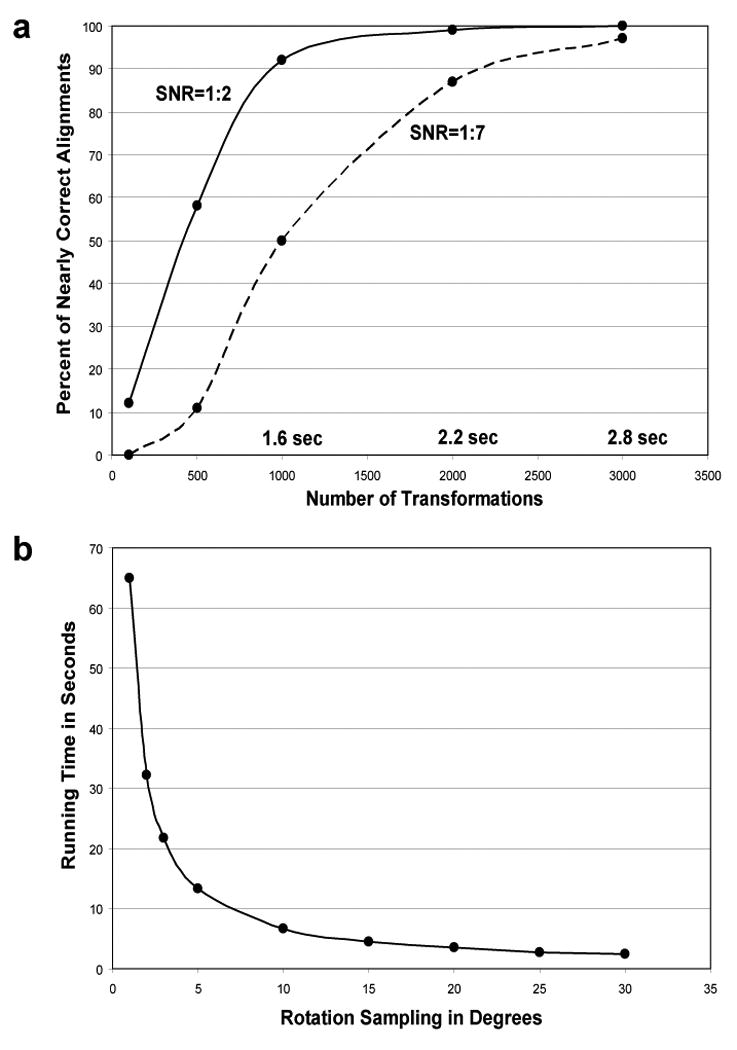
Running time measurements of FE+MI method and exhaustive transformation search implementation with MI as a scoring function. In both methods the translations are limited to at most 10 pixels from image center. (a) Transformation sampling accuracy and running time measurements for the FE+MI alignment method. The graph shows the percentage of nearly correct alignments, i.e., transformation sampling accuracy, of 50S ribosome images at SNR 1:2 and 1:7 with noise-free 70S ribosome templates. The accuracy is displayed as a function of the number of transformations employed after the pose-clustering procedure. The running time scales linearly with the number of transformations as each transformation should be evaluated with the scoring function. The running time of 2.2 seconds for 2000 transformations is equal to the time required for exhaustive rotational sampling at an angular step of 35 degrees. (b) Exhaustive search running time is displayed as a function of the size of angular steps used for rotational sampling.
