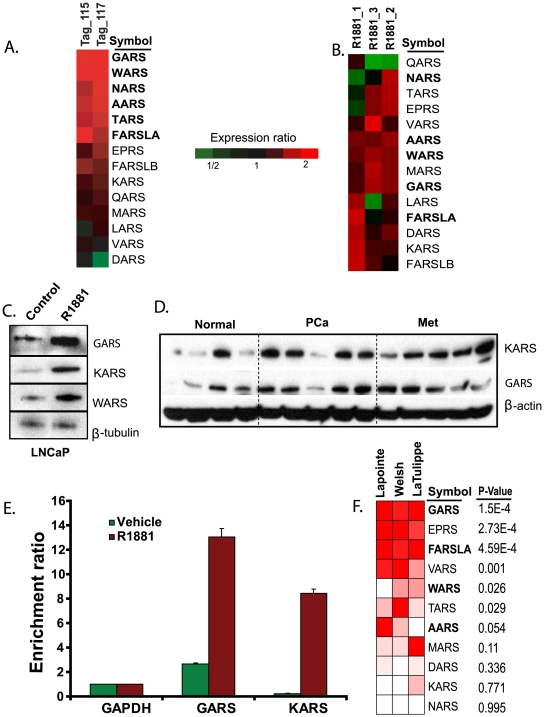
Figure 5. Androgen dependent up-regulation of aminoacyl tRNA synthetases in LNCaP, and their expression in prostate cancer.
A) Heat map showing the elevated expression of aminoacyl-tRNA synthetases identified from iTRAQ experiment. Only proteins in bold face are designated as androgen up-regulated in the iTRAQ data based on the threshold cut-off used. B) Heat map showing the concordant over-expression of transcripts measured using oligonucleotide microarray (Affymetrix U133 Plus 2.0) for aminoacyl-tRNA synthetases shown in A. C) Immunoblot analysis of aminoacyl t-RNA synthetases GARS, KARS, and WARS in response to androgen treatment in LNCaP cells. Beta-tubulin is used as loading control. D) Immunoblot analysis of KARS and GARS in localized prostate cancer (PCa), and metastatic prostate cancer (Met) compared to normal (benign) prostate tissues. Beta-actin is used as loading control. E) Promoter-occupancy of AR on aminoacyl tRNA genes. Chromatin immunoprecipitation (ChIP) -PCR analysis shows the androgen-dependent enrichment of AR-binding to the target promoters of GARS and KARS. Enrichment ratio was calculated based on the amount of target amplification against the input DNA, with primers for GAPDH promoter used as control. The primer sequences spanning the gene promoters are given in Table S5. F) Meta-analysis of the aminoacyl t-RNA synthetase genes (of proteins shown in A) across three prostate cancer gene expression profiling studies. Oncomine heat map view showing the over-expression of a subset of aminoacyl-tRNA synthetase genes in localized prostate cancer compared to their benign controls. Gene symbols of proteins that pass through the cut-off value and were assigned as androgen up-regulated in iTRAQ data are indicated in bold face. P-value represents the significance of expression in two of the three studies. Red, white, and blue (not present in the figure) indicates relative over, unchanged, and under expression, respectively.