Fig. 3.
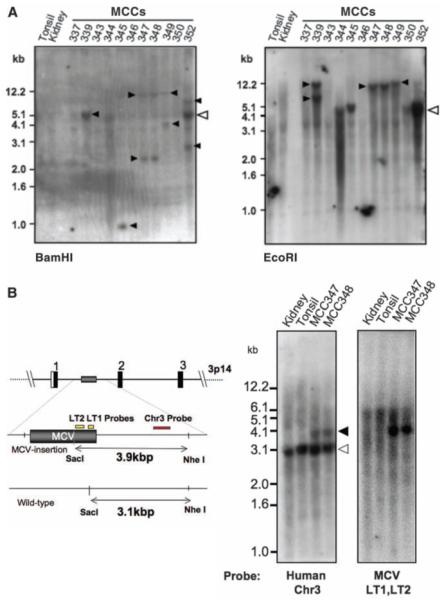
Clonal MCV integration in MCC tumors detected by direct Southern hybridization. (A) DNA digested with BamHI (left) or EcoRI (right) and Southern-blotted with MCV DNA probes reveals different banding patterns in each tumor, including >5.4-kb bands. Open arrowhead shows the expected position for MCV episomal or concatenated-integrated genome (5.4 kb) with corresponding bands present in tumors MCC344 and 350. Tumors MCC339, 345, 347, 348, and 349 have different band sizes and doublet bands (solid arrowheads), consistent with genomic monoclonal integration. MCC352 has a prominent 5.4-kb band as well as higher and lower molecular weight monoclonal integration bands (BamHI), consistent with an integrated concatemer. Tumors MCC337, 343, and 346 have no MCV DNA detected by Southern blotting [bands at 1.5 kb (kidney) and 1.2 kb (MCC346) are artifacts]. (B) Viral and cellular monoclonality in MCC347 and 348. Tumor MCC347 and its metastasis MCC348 were digested with SacI and NheI and Southern-blotted with unique human flanking sequence probe [Chr3 (red), left] or viral probes [LT1 and LT2 (yellow), right]. The wild-type human allele is present in all samples at 3.1 kb (left). The MCC tumors, however, have an additional 3.9-kb allelic band formed by MCV DNA insertion into chromosome 3p14. Hybridization with probes for MCV T antigen sequence (yellow, right) generates an identical band.
