Figure 5. Deletion and swap mutants establish the role of RRMs for phosphorylation initiation.
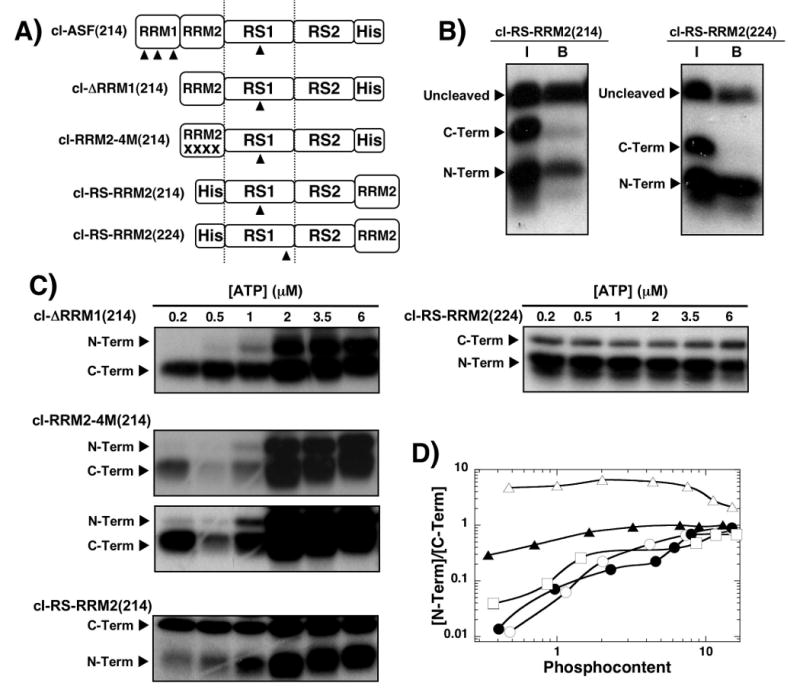
A) Deletion and swap mutants. Numbers in parentheses define the Arg-to-lys cleavage site in the RS domain. The same numbering scheme for the RS domain in wild-type ASF/SF2 is used for the swap and deletion mutants. B) Ni-resin pull-down assays. SRPK1 (1 μM) is used to phosphorylate 250 nM cl-RS-RRM2(214) and cl-RS-RRM2(224) before cleavage by LysC. Samples before binding to the N-resin (I) and after binding and washing (B) are displayed in the autoradiogram. C) ATP limitation experiments. Complexes of SRPK1 (1 μM) and mutants (250 nM) are phosphorylated for 20 min using varying 32P-ATP, cleaved with LysC and separated on SDS-PAGE. Two time exposures (20 & 70 min) for cl-RRM2-4M(214) gel were performed for better visualization of low and high ATP concentrations. D) N/C plot. The ratios of 32P in the N- and C-terminal fragments for cl-ΔRRM1(214) (●), cl-ASF(214) (○), cl-RRM2-4M(214) (□), cl-RS-RRM2(214) (▲), and cl-RS-RRM2(224) (△) are plotted against the total phosphoryl content
