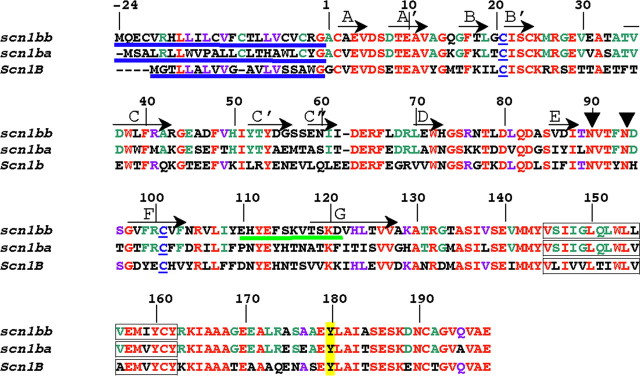
Figure 1.
Alignment of Scn1bb, Scn1ba_tv1, and Scn1b. Numbering of the amino acid residues in this figure corresponds to Scn1bb. Shown in red are the residues that are identical between Scn1bb, Scn1ba, and Scn1b. Residues in purple are residues that are shared by Scn1bb and Scn1b but not shared with Scn1ba. (Scn1ba comprises two different splice variants, Scn1ba_tv1 and Scn1ba_tv2. For purposes of clarity, we have shown the sequence for only one of the two splice variants, Scn1ba_tv1). Residues in green are conserved within the two fish paralogs but not shared with Scn1b. N-terminal signal peptides are underlined in blue. Also indicated in blue are the two cysteine residues in each subunit that are predicted to form the Ig loop domain. Predicted β-sheets in the Ig loop domain, based on the crystal structure of myelin P0 (Shapiro et al., 1996), are indicated by arrows and labeled with capital letters (A through G). Predicted glycosylation sites at asparagine residues in Scn1bb are indicated by arrowheads at N-90 and N-94. These sites were determined using NetNGlyc 1.0 (http://www.cbs.dtu.dk). Predicted transmembrane spanning segments are indicated as boxed residues. The tyrosine residue, Y-180, homologous to Y-181 in Scn1b (Malhotra et al., 2004) is highlighted in yellow. The peptide sequence used for anti-Scn1bb antibody production is underlined in green.