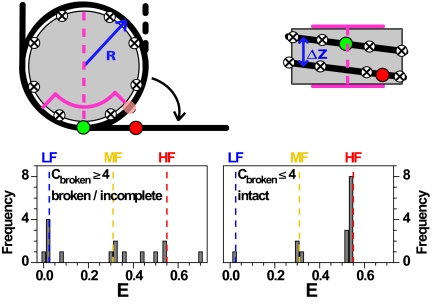
Fig. 5.
Geometric model for nucleosome unfolding. Nucleosomal DNA (black line) is wrapped around a cylindrical histone core (gray). Dyad (magenta dashed line), region with strong interactions (magenta line), contact points (circled crosses), donor and acceptor dyes (green and red spheres). In the top view, the semitransparent red circle represents the position of the acceptor in the fully wrapped nucleosome. The detached DNA segments were straight and tangential to the nucleosome core at the detachment point(s). Model parameters were estimated from the X-ray structure: Effective nucleosome radius R = 40 Å, rise per turn Δz = 45 Å, 80 bp per nucleosome turn, DNA length of 170 bp, donor dye position at 46.5 bp, acceptor dye position 136.5 bp, scaled Förster radius ßR0 = 61.2 Å with ß = 1.1 (for details see SI Appendix, section 2.7). (Lower) Possible FRET efficiency values from the geometrical model with contact points 10 bp apart, showing values obtained for the loss of ≤4 contact points (Right) and ≥4 contact points (Left).