Figure 1.
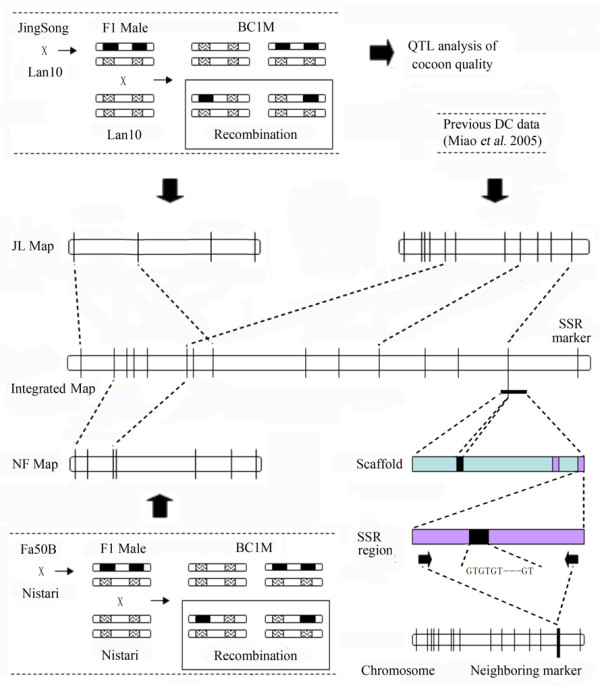
Outline of the study. We mapped SSR loci based on the mapping population Fa50B × Nistari and Jingsong × Lan10. Two new SSR linkage maps (NF and JL) were constructed, and the resultant data set was merged with previous data sets to generate the integrated linkage map. The mapped SSR sites were used to localize silkworm genomic scaffolds and SSRs were sought in the extended genomic sequences to develop neighboring markers. Thus, informative bins were formed by integrating the substitutes with the mapped sites.
