Figure 3.
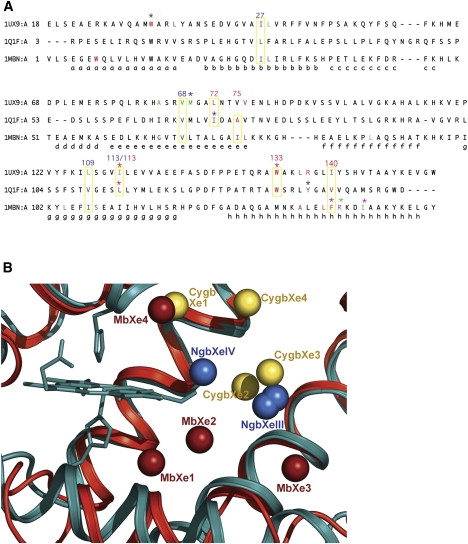
Structural superposition of Ngb•Xe with swMb•Xe and hCygb•Xe. (A) Structure-based sequence alignments were performed using CE (46). Sequences are reported with their relative PDB code (1UX9 for Cygb•Xe; 1Q1F for Ngb; and 1MBN for swMb), and the α-helices are labeled according to the globin fold topology rules. Amino acids that form a xenon site are color coded green (Xe1), magenta (Xe2), red (Xe3 and XeIII), and blue (Xe4 and XeIV). Amino acids that participate at two sites are treated with the same color code but have an asterisk above them. The alignments show that some residues belonging to the NgbXeIII site (red; I72, A75, L113, W133, and V140) or the NgbXeIV site (blue; L27, V68, V109, and L113) are conserved or have the same chemical nature with respect to the amino acids that form the Xe sites of Cygb and/or Mb. We enclose these residues in yellow boxes, also indicating their position in Ngb. (B) Comparison of the Xe atom positions in Ngb (blue), Cygb (yellow), and Mb (red) after superposition of the protein main chain only. The Xe atoms are represented as spheres. Cygb and the heme group of Mb are not shown. Distances between Xe atoms are reported in Table 2. Xe proximal sites are present only in Mb.
