A regulator of G protein signaling, RGS2, moonlights in protein synthesis control.
Abstract
The regulator of G protein signaling (RGS) proteins are a family of guanosine triphosphatase (GTPase)–accelerating proteins. We have discovered a novel function for RGS2 in the control of protein synthesis. RGS2 was found to bind to eIF2Bϵ (eukaryotic initiation factor 2B ϵ subunit) and inhibit the translation of messenger RNA (mRNA) into new protein. This effect was not observed for other RGS proteins tested. This novel function of RGS2 is distinct from its ability to regulate G protein–mediated signals and maps to a stretch of 37 amino acid residues within its conserved RGS domain. Moreover, RGS2 was capable of interfering with the eIF2–eIF2B GTPase cycle, which is a requisite step for the initiation of mRNA translation. Collectively, this study has identified a novel role for RGS2 in the control of protein synthesis that is independent of its established RGS domain function.
Introduction
The regulator of G protein signaling (RGS) and RGS-like proteins comprise a family of at least 30 members that have been grouped into five subfamilies based on their structural and sequence homologies (Ross and Wilkie, 2000). RGS proteins possess a conserved ∼120–amino acid RGS domain that promotes their association with heterotrimeric G protein α subunits and confers their function as GTPase-accelerating proteins (GAPs). RGS2 belongs to the B/R4 subfamily of RGS proteins that are characterized by relatively simple structures wherein an RGS domain is flanked by short amino and carboxyl termini. Although RGS2 does not appear to contain any of the other established protein-interacting domains (e.g., PSD-95/Dlg/zona occludens-1, ras binding domain, and GoLoco) that have been identified within some RGS protein subfamilies, accumulating evidence supports the hypothesis that RGS2 may regulate cellular activity in a manner that is distinct from its known RGS domain function (Sinnarajah et al., 2001; Salim et al., 2003; Roy et al., 2006a; Schoeber et al., 2006). For example, it has recently been shown that RGS2 interacts with the TRPV6 member of the transient receptor potential family of cation channels to disrupt both Na+ and Ca2+ currents (Schoeber et al., 2006). This effect was dependent on a stretch of amino acids (1–82) situated outside of the RGS domain, as it was not observed with an amino-terminal truncation mutant. RGS2 also binds to tubulin to enhance microtubule polymerization, and this effect is dependent on a stretch of 20 amino acids located outside of its RGS domain (Heo et al., 2006). These studies support additional functions for RGS2 other than its role as a GAP for Gαq (Ingi et al., 1998; Bernstein et al., 2004) and regulator of Gαs-mediated adenylyl cyclase activation (Roy et al., 2006a). In this study, we present evidence for a novel and unexpected function wherein RGS2 inhibits the translation of mRNA into protein by blocking the activity of the rate-controlling eIF2B.
Dysregulation of the protein synthesis machinery can contribute to human disease states such as cancer, diabetes, cardiac hypertrophy, and neurodegeneration (Proud, 2007). The stages of mRNA translation are initiation, elongation, and termination. In eukaryotes, cellular control of initiation is governed by a family of proteins referred to as eIFs (eukaryotic initiation factors; Proud, 2005). The rate-limiting step in translation occurs during initiation and is regulated by the heterotrimeric (αβγ) GTPase eIF2 (Kimball, 1999). In its activated conformation, eIF2-GTP can form a ternary complex with Met-tRNAi, which then binds to the 40S ribosomal subunit to initiate protein synthesis. The guanine nucleotide–bound state of eIF2 is itself governed by the heteropentameric (αβδϵγ) guanine nucleotide exchange factor (GEF), eIF2B, and the eIF2-specific GAP, eIF5. One of the mechanisms to regulate eIF2 activity is phosphorylation of its α subunit in response to various cellular stressors. This is mediated by four stress-activated kinases: haem-regulated inhibitor, general control nonderepressible-2, protein kinase activated by double-stranded RNA, and pancreatic endoplasmic reticulum eIF2α kinase (Dever, 2002; Ron, 2002). Phosphorylation of this highly conserved serine acts to decrease the dissociation rate of eIF2 from eIF2B, which essentially converts eIF2 from a substrate into a competitive inhibitor of eIF2B GEF activity. However, the control of protein synthesis in response to stress is multifaceted and cannot be solely explained by phosphorylation events.
Using a yeast two-hybrid screen, we have identified an interaction between RGS2 and eIF2Bϵ. This association between RGS2 and eIF2Bϵ was also observed between the endogenous proteins in cells. RGS2 was capable of inhibiting de novo protein synthesis in a manner independent of its RGS domain function. Collectively, this work has identified and characterized RGS2 as a novel component in the control of protein synthesis.
Results
Physical interaction between RGS2 and eIF2Bϵ
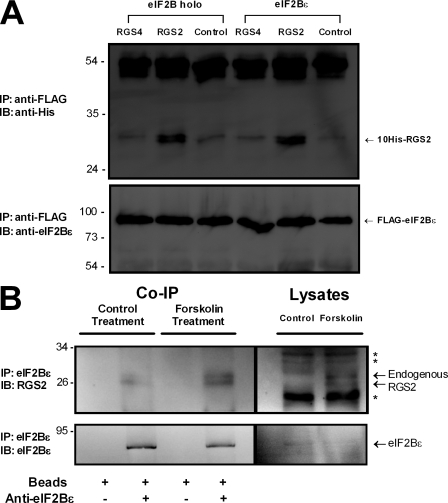
Initial experiments using full-length human RGS2 as bait in a yeast two-hybrid screen pointed to an interaction with eIF2Bϵ (4 of 21 total positives). In contrast, RGS4 was not observed to interact with eIF2Bϵ when screened against the same mouse brain cDNA library. The 45–amino acid sequence identified in the RGS2 screen matches perfectly with amino acid residues 550–594 of mouse eIF2Bϵ (equivalent to 554–598 in human). Notably, this corresponds to the highly conserved catalytic surface of eIF2Bϵ that enables the heteropentameric eIF2B complex to promote GDP dissociation from eIF2 (Boesen et al., 2004; Mohammad-Qureshi et al., 2007). It follows that if RGS2 were to bind to this region of eIF2Bϵ, it could alter the ability of eIF2B to promote protein synthesis. We first sought to confirm whether the interaction between RGS2 and eIF2Bϵ could occur at the protein level. We mixed purified RGS2 or RGS4 with lysates from Sf9 cells overexpressing eIF2Bϵ and examined whether there was an association between the proteins. Consistent with the results of the yeast two-hybrid screen, RGS2 interacted with eIF2Bϵ whether it was expressed as a monomer or as part of the pentameric eIF2B complex (i.e., ± eIF2Bα/eIF2Bβ/eIF2Bδ/eIF2Bγ; Fig. 1 A). Thus, it appears that the presence of the other four subunits does not sterically hinder the ability of eIF2Bϵ to associate with RGS2. In contrast, RGS4 did not specifically associate with either the eIF2B pentamer or monomeric eIF2Bϵ (Fig. 1 A). Next, we investigated whether there was an association between endogenously expressed RGS2 and eIF2Bϵ; therefore, we addressed whether the interaction could occur between the native proteins in UMR-106 osteoblast-like cells. For these experiments, we were required to up-regulate endogenous RGS2 expression by way of the diterpene forskolin, given that native RGS2 in osteoblast cultures is undetectable by immunoblot analysis under basal conditions (Roy et al., 2006b). Endogenous eIF2Bϵ was immunoprecipitated from whole cell lysates, and the immune complex was examined for RGS2 immunoreactivity. Indeed, endogenously expressed RGS2 was observed to coimmunoprecipitate with eIF2Bϵ (Fig. 1 B). RGS2 expression in forskolin-treated cells and from the coimmunoprecipitation assay presented as a doublet band, which is consistent with a recent report that RGS2 can have multiple protein products resulting from alternative translation initiation sites (Gu et al., 2008). This observed association between RGS2 and eIF2Bϵ in lysates from nontransfected cells shows that the interaction between the two proteins is genuine and thus implies a possible role for endogenous RGS2 in mRNA translation.
Figure 1.
In vitro interaction between RGS2 and eIF2Bϵ. (A) Flag-tagged eIF2Bϵ was expressed in Sf9 cells in either its monomeric form or as part of the eIF2B holoprotein (i.e., ± eIF2Bα/eIF2Bβ/eIF2Bδ/eIF2Bγ). The precleared cell lysate was obtained as described in Materials and methods, and the indicated RGS protein was added to these lysates to a final concentration of 500 nM. The mixture was immunoprecipitated with anti-Flag antibody, and samples were run on SDS-PAGE and transferred. Membranes were probed with antihistidine antibody to identify coimmunoprecipitated RGS proteins (top) or anti-eIF2Bϵ to view immunoprecipitated eIF2Bϵ (bottom) from lysate. The blots shown are representative of at least three independent experiments. (B) Coimmunoprecipitation of endogenous RGS2 with endogenous eIF2Bϵ. UMR-106 osteoblast-like osteosarcoma cells were treated for 3 h with 100 µM forskolin to induce RGS2 expression. Vehicle control and forskolin-treated cell lysates (1 mg total protein) were incubated with protein A/G agarose beads either without (lanes 1 and 3) or with (lanes 2 and 4) anti-eIF2Bϵ antibody, or else were added directly to the gel (100 µg total protein; lanes 5 and 6). Protein A/G agarose beads were extensively washed, and the bound protein was removed by heating and added to the gels (lane 1–4). The SDS-PAGE gel was transferred to PVDF membrane and probed using anti-RGS2 antibody (top) and stripped and reprobed using the same eIF2Bϵ antibody used for immunoprecipitation (IP; bottom). The results shown are typical of three independent experiments. Asterisks denote nonspecific immunoreactive bands. IB, immunoblot. Molecular mass indicators are expressed in kilodaltons.
Inhibition of protein synthesis by RGS2
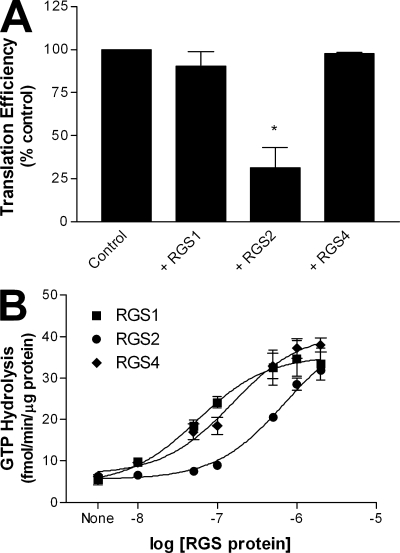
The role of eIF2B in the control of protein synthesis is to promote guanine nucleotide exchange on the eIF2γ subunit. Protein translation cannot be initiated in the absence of a functional eIF2–eIF2B relationship. Thus, we hypothesized that the interaction between RGS2 and eIF2Bϵ may act to interfere with de novo protein synthesis. Therefore, we monitored the production of the luminescent protein, Coleoptera luciferase, from its mRNA using a rabbit reticulocyte lysate-based in vitro translation assay. Addition of RGS2 resulted in a significant decrease in protein synthesis compared with control (Fig. 2 A). In contrast, neither RGS1 (despite its reported interaction with eIF3δ; see Discussion) nor RGS4 had any effect on protein synthesis (Fig. 2 A). Each of the three purified RGS proteins was functional with respect to its known GAP activity, as all were able to dose-dependently increase the rate of M1 muscarinic receptor–stimulated G11 GTP hydrolysis to approximately the same extent (Fig. 2 B).
Figure 2.
Inhibition of protein synthesis by RGS2. (A) The effects of 4 µM purified RGS proteins on the synthesis of the reference luciferase protein were examined in a reticulocyte-based in vitro translation assay as described in Materials and methods. (B) Concentration response effects of full-length RGS proteins on steady-state, agonist-stimulated GTPase activity of M1 muscarinic receptor–activated Gα11 as described in Materials and methods. The data are presented as mean ± SEM of three independent experiments performed in triplicate. *, P < 0.05 versus control (one-sample t test).
RGS2 regulation of protein synthesis and the RGS domain
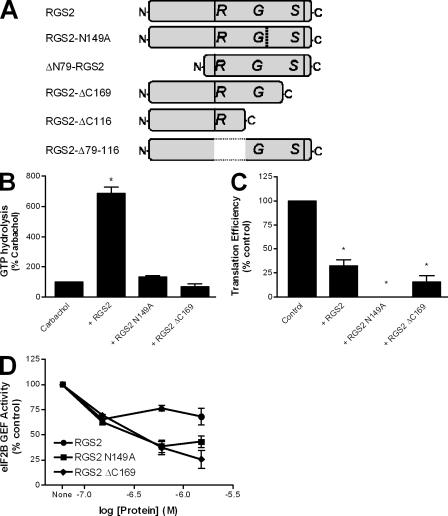
To better define the mechanism by which RGS2 controls protein synthesis, we examined the role of its RGS domain. We generated two mutations within this region of RGS2 that would act to decrease or eliminate its GTPase-accelerating function: RGS2-N149A and RGS2-ΔC169 (Fig. 3 A). The N149A point mutation is thought to disrupt a critical contact point between RGS2 and G protein α subunits, whereas ΔC169 removes a substantial portion of the RGS domain at its carboxyl terminus. As expected, neither of these RGS2 mutants was able to increase agonist-stimulated GTP hydrolysis above that of agonist alone (Fig. 3 B). In contrast, when the two RGS2 mutants were used in the in vitro translation assay, both proteins retained the ability to inhibit the translation of luciferase mRNA into protein to the same degree as that of wild-type RGS2 (Fig. 3 C). These results imply that the ability of RGS2 to inhibit protein synthesis is independent of its effects on G proteins.
Figure 3.
RGS2-mediated inhibition of protein synthesis is independent of its RGS domain function. (A) Schematic of the purified RGS2 proteins used throughout the study. (B and C) The effects of full-length RGS2, RGS2-N149A, and RGS2-ΔC169 (4 µM) were examined on steady-state, agonist-stimulated GTPase activity of M1 muscarinic receptor–activated Gα11 (B) or synthesis of the reference luciferase protein in a reticulocyte-based in vitro translation assay (C) as described in Materials and methods. The data are presented as mean ± SEM of three independent experiments performed in triplicate. *, P < 0.05 versus control (one-sample t test). (D) The ability of 150 nM purified eIF2B to promote dissociation of [3H]GDP from purified eIF2 was examined as described in Materials and methods. This activity was assessed in the absence and presence of three concentrations of RGS2, RGS2N149A, and RGS2ΔC169. The data are presented as mean ± SEM of four independent experiments performed in duplicate.
We hypothesized that the binding of RGS2 to eIF2Bϵ might disrupt the eIF2–eIF2B GTP cycle. Thus, we examined the effects of RGS2 on the GEF activity of eIF2B by measuring the potential of eIF2B to promote the dissociation of [3H]GDP from eIF2. As illustrated, all three of the RGS2 proteins examined were able to dose-dependently inhibit eIF2B GEF activity for eIF2 (Fig. 3 D). Collectively, these data show that RGS2 can interfere with eIF2B GEF activity in a manner independent of its established G protein effects.
Identification of the RGS2 domain involved in regulating protein synthesis
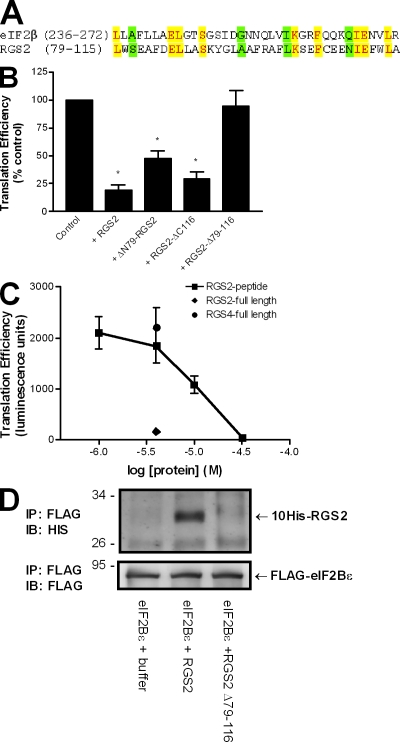
The observation that RGS2 can interfere with the eIF2–eIF2B GTP cycle suggests that RGS2 may bind to eIF2Bϵ in a manner similar to that of eIF2β. We identified a stretch of 37 amino acid residues within the RGS2 protein sequence (79–115) that shared 35% homology (nine identical and four conserved) with the eIF2Bϵ-binding domain of eIF2β (200–333; Fig. 4 A; Kimball et al., 1998). This corresponding sequence within other RGS and RGS-like proteins did not exhibit the same degree of identity. Structural analyses revealed that the homologous regions of human RGS2 (Protein Data Bank accession no. 2af0; Soundararajan et al., 2008) and bacterial aIF2β (Protein Data Bank accession no. 1nee; Gutiérrez et al., 2004) were each comprised of two α helices oriented at a 45° angle and connected by a short linker. We used this homologous sequence of 37 amino acids as a point of reference to generate ΔN79-RGS2, RGS2-ΔC116, and RGS2-Δ79–116 (Fig. 3 A) in an attempt to identify the active domains within RGS2 that confer its novel function. Deletion of the RGS2 amino terminus did not interfere with RGS2 GAP activity. As reported previously (Bernstein et al., 2004), both wild-type RGS2 and ΔN79-RGS2 were able to promote agonist-stimulated Gα11 GTP hydrolysis (unpublished data). This is consistent with the fact that truncation of RGS2 upstream of residue 79 leaves the RGS box intact. However, RGS2-ΔC116 and RGS2-Δ79–116 were without RGS2 GAP activity (unpublished data). This was expected given that these mutations disrupt the integrity of the RGS domain. The three RGS2 deletion mutants were used in the translation assay to examine their effects on protein synthesis. Truncation of RGS2 either upstream or downstream of the homologous 37–amino acid sequence did not disrupt the ability of RGS2 to impair protein synthesis. RGS2-ΔC116 and ΔN79-RGS2 each inhibited mRNA translation to a comparable level as that of wild-type RGS2 (Fig. 4 B). In contrast, RGS2-Δ79–116 had no effect on protein synthesis (Fig. 4 B) and correspondingly failed to interact with monomeric eIF2Bϵ (Fig. 4 D). Additional experiments assessed the effect of a purified RGS2 peptide corresponding to residues 79–116 on translation. The results revealed that the RGS2 peptide was capable of inhibiting the translation of luciferase mRNA into protein in a concentration-dependent manner (Fig. 4 C). Collectively, these data demonstrate that RGS2 amino acids 79–116 are both sufficient and necessary to inhibit protein synthesis.
Figure 4.
RGS2-mediated inhibition of protein synthesis and binding to eIF2Bϵ is dependent on amino acids 79–116. (A) Comparison between the putative eIF2Bϵ-interacting domain of RGS2 and the established eIF2Bϵ-interacting domain of eIF2β. Identical residues are denoted by yellow shading, whereas conserved substitutions are indicated in green. (B) The effect of full-length RGS2, ΔN79-RGS2, RGS2-ΔC116, and RGS2-Δ79–116 (4 µM) was examined on luciferase protein synthesis in a reticulocyte-based in vitro translation assay as described in Materials and methods. The data are presented as mean ± SEM of three independent experiments performed in triplicate. *, P < 0.05 versus control (one-sample t test). (C) The effect of RGS2, RGS4, or an RGS2 peptide corresponding to amino acid residues 79–116 on luciferase protein synthesis. The data are presented as mean ± SEM of three independent experiments performed in duplicate. (D) RGS2 and RGS2-Δ79–116 were examined for interactions with monomeric eIF2Bϵ as described for Fig. 1. The blots are representative of three independent experiments. IB, immunoblot. Molecular mass indicators are expressed in kilodaltons.
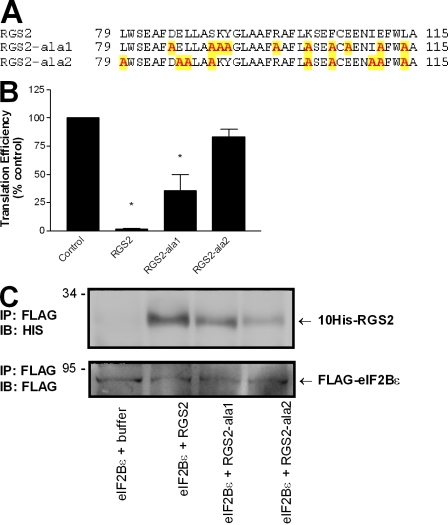
We went on to generate two additional RGS2 mutants termed RGS2-ala1 and RGS2-ala2 in an attempt to further characterize this new domain of RGS2 that is involved in protein translation (Fig. 5 A). The RGS2-ala mutants each contain a series of alanine substitutions at strategic locations between residues 79 and 116. For RGS2-ala1, the substitutions are based on the charge and position of the native residue within the helix–linker–helix structure of wild-type RGS2, whereas the substitutions in RGS2-ala2 correspond to the conserved residues between the homologous segments of eIF2β and RGS2 (Fig. 4 A). RGS2-ala1 and RGS2-ala2 were examined for their potential to interact with eIF2Bϵ and regulate de novo protein synthesis. In comparison with wild-type protein, RGS2-ala2 had minimal interactions with monomeric eIF2Bϵ and, accordingly, did not have a significant impact on the translation of luciferase mRNA (Fig. 5). In contrast, RGS2-ala1 retained binding to eIF2Bϵ and also significantly decreased protein translation (Fig. 5). These results further reinforce the proposal that this segment of RGS2 is involved in binding eIF2Bϵ in the regulation of protein synthesis and suggest that the common residues between eIF2β and RGS2 may contribute to eIF2Bϵ binding.
Figure 5.
RGS2 residues involved in binding eIF2Bϵ and regulating protein synthesis. (A) Identity and location of RGS2 amino acids targeted for alanine substitution (yellow shading). (B) The effect of RGS2, RGS2-ala1, and RGS2-ala2 on luciferase protein synthesis as described in Materials and methods. The data are presented as mean ± SEM of three independent experiments performed in duplicate. *, P < 0.05 versus control (one-sample t test). (C) Interactions between monomeric eIF2Bϵ and wild-type RGS2, RGS2-ala1, and RGS2-ala2. The blots are representative of two independent experiments. IB, immunoblot. Molecular mass indicators are expressed in kilodaltons.
RGS2 residues 79–116 inhibit protein synthesis in cells
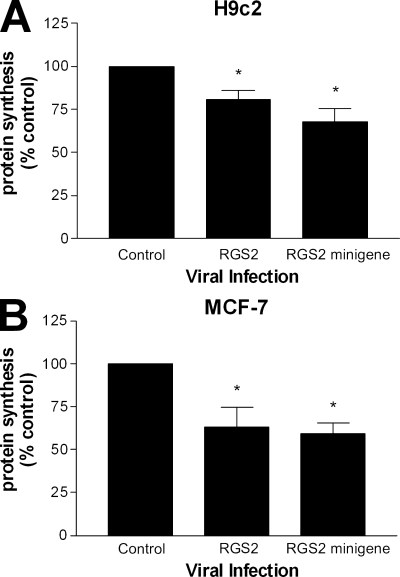
We next assessed whether RGS2 and its putative eIF2Bϵ-interacting domain had any impact on protein synthesis in cells. We used two cellular models for these experiments, H9c2 and MCF-7 cells, and infected them with adenovirus coding for full-length RGS2 and an RGS2 minigene comprising residues 79–116. In both of the cell lines tested, RGS2 and the RGS2 minigene were able to attenuate de novo protein synthesis to comparable levels (Fig. 6). The magnitude of inhibition was not as robust as that observed in the in vitro translation assay, but this was not surprising given the complexity of the protein synthesis machinery in a whole cell context.
Figure 6.
RGS2 amino acids 79–116 are sufficient to inhibit cellular protein synthesis. (A and B) H9c2 cardiomyoblasts (A) and MCF-7 adenocarcinoma cells (B) were infected with adenovirus (MOI = 4) encoding GFP (control), RGS2, or an RGS2 minigene for 48 h and used in a [3H]leucine protein synthesis assay as described in Materials and methods. The data were calculated as cpm/µg of total protein, normalized as a percentage of control, and are expressed as the mean ± SEM of five independent experiments performed in triplicate. *, P < 0.05 versus control (one-sample t test).
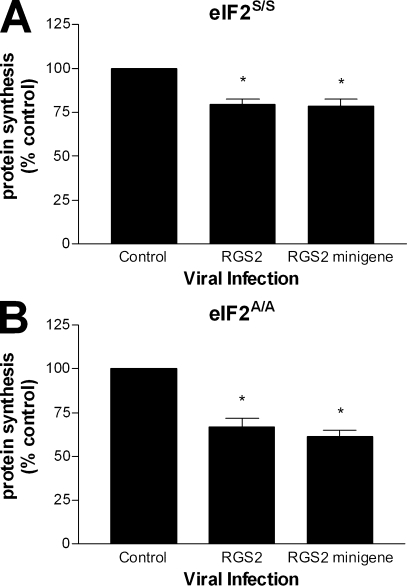
One possible contributing factor to the observed RGS2 effect on cellular protein synthesis may be that viral infection could promote phosphorylation of serine 51 on eIF2α, which itself may impede global protein synthesis. To address this issue, we made use of a previously described mouse embryonic fibroblast (MEF) cell line in which the gene for eIF2α was targeted to mutate the serine 51 phosphorylation site to a nonphosphorylatable alanine residue (Scheuner et al., 2001). Control MEF cells (eIF2S/S) and mutant MEF cells (eIF2A/A) were used to measure de novo protein synthesis after infection with RGS2 and the RGS2 minigene (Fig. 7). Consistent with our results using the H9c2 and MCF-7 cells, a decrease in protein synthesis was also observed in both of these MEF cell lines. The fact that the decrease in de novo protein synthesis was retained in the eIF2A/A cells argues that the mechanism by which RGS2 inhibits translation is independent of serine 51 phosphorylation on eIF2α. There were no differences observed in the cellular uptake of radiolabeled leucine between the various infection conditions (unpublished data). These data further reinforce the notion that RGS2 may serve a regulatory role in protein synthesis through its interactions with eIF2Bϵ.
Figure 7.
RGS2-mediated inhibition of cellular protein synthesis is independent of eIF2α serine 51 phosphorylation. (A and B) MEF eIF2S/S (A) and eIF2A/A (B) cell lines were infected with adenovirus (MOI = 4) encoding RGS4 (control), RGS2, or an RGS2 minigene for 48 h and used in a [3H]leucine protein synthesis assay as described in Materials and methods. The data were calculated as cpm/µg of total protein, normalized as a percentage of control, and are expressed as the mean ± SEM of three independent experiments performed in triplicate. *, P < 0.05 versus control (one-sample t test).
Protein synthesis is elevated in the absence of endogenous RGS2
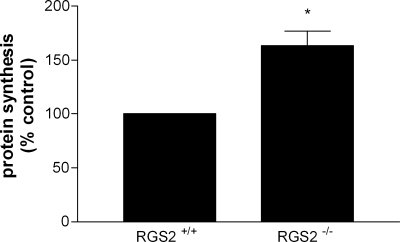
Lastly, we went on to investigate the effect of silencing RGS2 expression on cellular protein synthesis. We isolated hepatocytes from wild-type and RGS2 knockout mice (RGS2−/−) and compared their levels of de novo protein synthesis. These experiments revealed a 60% increase in protein synthesis in RGS2−/− cells compared with wild-type controls (Fig. 8). These results are in line with previous observations that knocking out either general control nonderepressible-2 or haem-regulated inhibitor decreases eIF2α phosphorylation and results in elevated levels of protein synthesis (Han et al., 2001; Lu et al., 2001; Deng et al., 2002). We did not observe any difference in the rate of protein catabolism between the two hepatocyte cell cultures (unpublished data). These results are consistent with the notion that in the absence of endogenous RGS2, there would be fewer restrictions on the translational machinery and, therefore, elevated levels of protein synthesis. Collectively, this study provides strong evidence that RGS2 plays a pivotal role in regulating the protein synthesis machinery.
Figure 8.
Protein synthesis is elevated in RGS2−/− mice. Hepatocytes from wild-type control and RGS2−/− mice were prepared and used in a [3H]leucine protein synthesis assay as described in Materials and methods. The data were calculated as cpm/µg of total protein, normalized as a percentage of control, and are expressed as the mean ± SEM of three independent experiments performed in triplicate. *, P < 0.05 versus control (paired t test).
Discussion
These results outline a novel function for RGS2 in the control of protein synthesis in addition to its established role as a GAP for heterotrimeric G proteins. RGS2 is able to bind to the ϵ subunit of the GEF, eIF2B, and this interaction appears to interfere with the initiation of mRNA translation by preventing guanine nucleotide exchange on eIF2. The notable similarities in protein sequence and tertiary structure between the homologous regions of RGS2 and eIF2β led to the hypothesis that RGS2 residues 79–116 play a role in binding eIF2Bϵ. Mutagenesis analyses confirmed that this segment of RGS2 is required for its interaction with eIF2Bϵ and regulation of translation. Furthermore, using an alanine substitution approach, we were able to identify key residues involved in the protein–protein interaction. The near complete loss of eIF2Bϵ binding and inhibition of in vitro translation by RGS2-ala2 suggests the nine substituted residues within this mutant protein are critical contact points for RGS2–eIF2Bϵ interactions. This is consistent with the fact that these nine residues are identical between the homologous segments of RGS2 and eIF2β. It is noteworthy to point out the four amino acid substitutions that are unique to RGS2-ala2: L79, E86, L87, and I110. This suggests that these residues are responsible for the observed difference in eIF2Bϵ binding between RGS2-ala2 and RGS2-ala1; the residual interactive forces for binding eIF2Bϵ would presumably be made up of the remaining five overlapping substitutions, although this cannot be conclusively stated at this point in time. The successful crystallization of an RGS2–eIF2Bϵ complex (or parts thereof) will provide more information on the molecular determinants for this interaction.
Several potential links between G protein–mediated signaling networks and cellular control of protein synthesis have previously been noted, although no direct effects on the initiation of mRNA translation have been established. The α subunit of eIF2B has been reported to interact with the carboxyl tails of the α2A, α2B, α2C, and β2 adrenergic receptors in a yeast two-hybrid screen, but not to the carboxyl tail of the vasopressin receptor (Klein et al., 1997). Similarly, eIF2Bα has been shown to interact with the third intracellular loop of the α2B and α2C adrenergic receptors in a manner that may be dependent on 14–3-3ζ (Prezeau et al., 1999). The third intracellular loop of the M4 muscarinic receptor, but not M1 or M2, was also reported to associate with eukaryotic elongation factor 1A2 to promote guanine nucleotide exchange on the latter (McClatchy et al., 2002), which was suggested to be a mechanism of regulating M4 muscarinic receptor recycling (McClatchy et al., 2006). No effects of RGS proteins on mRNA translation have been reported until now, although RGS1 was identified as a binding partner for eIF3δ in a yeast two-hybrid screen (http://www.signaling-gateway.org/data/Y2H/cgi-bin/y2h_int.cgi?id=17628). The latter interaction was not confirmed at the protein level, and our study has not shown RGS1 to have any effect on protein synthesis. Nonetheless, these data support a role for G protein–mediated signaling networks in the control of protein synthesis.
Under conditions of cellular stress such as oxidative damage, nutrient deprivation, viruses, and heat shock, the overall rate of mRNA translation into protein is reduced (although the synthesis of stress-related proteins is maintained or increased through specialized alternative pathways; Ron, 2002; Wek et al., 2006). A variety of mechanisms exist to reduce global protein synthesis, and for the most part, these involve changes in initiation. Of particular importance is the rate-limiting eIF2–eIF2B interaction, which depends upon both the activity and the availability of the exchange factor eIF2B. The best characterized inhibitory mechanism involves the phosphorylation of Ser51 on the eIF2α subunit (Gebauer and Hentze, 2004). Other mechanisms for inhibiting mRNA translation by way of eIF2B have also been reported. For example, GSK3 directly phosphorylates eIF2Bϵ at serine 540 in intact cells, and this inhibits eIF2B GEF activity by up to 80% (Welsh et al., 1998). In addition, it has been shown that the effects of eIF2B on eIF2 can be decreased by the binding of the latter to eIF5. The primary function of eIF5 is to promote GTP hydrolysis by eIF2; however, when present at elevated levels, eIF5 may act to sequester eIF2 from eIF2B, thereby impeding protein synthesis (Singh et al., 2006). These results suggest a comparable inhibitory mechanism wherein the interaction between eIF2 and eIF2B similarly is hindered by the binding of a third protein, in this case the association of RGS2 with eIF2B. Although the specific protein target differs, the functional consequence in both cases is an attenuation of guanine nucleotide exchange on eIF2.
One interesting aspect of RGS2 signaling is that RGS2 mRNA and protein expression can be up-regulated in response to the same forms of cellular stress that inhibit protein synthesis, including oxidative stress, heat shock, DNA damage, and mechanical stress (Siderovski et al., 1994; Zmijewski et al., 2001; Song and Jope, 2006; Santos de Araujo et al., 2007). For example, treatment of 1321N1 astrocytoma cells with H2O2 leads to the up-regulation of RGS2 mRNA and protein in a concentration-dependent manner, and subjecting these cells to heat shock also results in increased levels of RGS2 mRNA in a time-dependent manner (Zmijewski et al., 2001). Likewise, when SH-SY5Y cells were treated with the DNA-damaging agent camptothecin, RGS2 mRNA levels were found to increase, whereas RGS4 mRNA levels decreased (Song and Jope, 2006). Although eIF2α phosphorylation and RGS2 up-regulation tend to be triggered by similar stimuli, these two processes are likely distinct and separated on a temporal level. It is conceivable that changes in translation mediated by stress-activated kinases could be prolonged by RGS2 because the increase in its protein levels might be expected to coincide with the eventual dephosphorylation of serine 51 in eIF2α. It still remains to be demonstrated whether endogenous RGS2 can attain sufficient protein levels in cells to replicate the effects observed on de novo protein synthesis after viral infection. This has been hindered primarily by the fact that the majority of agents and stressors that up-regulate endogenous RGS2 also induce eIF2α phosphorylation, thereby making it difficult to assign the contribution from each signaling event toward controlling protein synthesis. However, the observation that levels of RGS2 required for maximal GAP activity (its established biochemical function) closely mirror those needed in the in vitro translation assay suggest that endogenous levels of RGS2 are sufficient to regulate translation in cells. Moreover, we have shown that endogenous RGS2 and eIF2Bϵ can be coimmunoprecipitated. Other factors, such as the complex and changeable intracellular distribution patterns found with RGS2 and the potential effects of posttranslational modifications, may also play a role. Our attempts to circumvent these obstacles by using the RGS2−/− mice provide strong evidence that endogenous levels of RGS2 are sufficient to regulate translation; however, we cannot rule out the possibility that the observed difference in protein synthesis between wild-type and RGS2−/− cells is not the result of an indirect effect unrelated to RGS2 regulation of eIF2Bϵ.
Its up-regulation in response to stress suggests that RGS2 may help to maintain cellular integrity. Indeed, RGS2 has previously been shown to act as a negative regulator of α1-adrenergic receptor–stimulated cardiomyocyte hypertrophy (Zou et al., 2006). In primary cultures of ventricular myocytes, induction of hypertrophy by the α1-adrenergic receptor agonist phenylephrine resulted in the selective increase of RGS2 mRNA over RGS1, RGS3, RGS4, and RGS5 (Zou et al., 2006). Overexpression of RGS2 in these cells completely blocked the phenylephrine-dependent increase in cell size as well as the induced expression of various genetic markers for cardiac hypertrophy (Zou et al., 2006). These results are consistent with the report that down-regulation of endogenous RGS2 by way of RNAi exacerbates cardiomyocyte hypertrophy (Zhang et al., 2006). Such findings are typically thought to reflect the negative effects of RGS2 on Gq-mediated signaling; however, given that hypertrophy is characterized by an increase in protein synthesis and cell size, one could argue based on these results that stress-induced up-regulation of RGS2 expression may impede the development of cardiac hypertrophy by inhibiting global protein synthesis. Indeed, de novo protein synthesis appears to be a requirement for cardiomyocyte hypertrophy to develop in some cell-based models (Hardt et al., 2004; Shan et al., 2007).
In conclusion, the present study has identified RGS2 as a component of the protein synthesis machinery. This novel function of RGS2 may serve as an unexpected regulatory mechanism of the cellular stress response. Furthermore, the observation that a minigene coding for the RGS2 residues involved in binding eIF2Bϵ (79–116) is sufficient to inhibit de novo protein synthesis provides a template for the development of drugs to treat diseases that are characterized by impaired protein synthesis such as neoplasias, diabetes, cardiac hypertrophy, and neurodegeneration (Proud, 2007).
Materials and methods
Reagents and clones
Plasmids encoding histidine-tagged wild-type RGS2, RGS4, and ΔN79-RGS2 were provided by J. Hepler (Emory University, Atlanta, GA). The cDNA for RGS1 was provided by D. Siderovski (University of North Carolina at Chapel Hill, Chapel Hill, NC) and subcloned into pET19b. The baculovirus coding for an M1 muscarinic receptor–G11 fusion protein was provided by T. Haga (University of Tokyo, Hongo, Japan). Baculoviruses encoding all five Flag-tagged subunits of eIF2B were generated as described previously (Fabian et al., 1997). RGS2-N149A, RGS2-ΔC116, and RGS2-ΔC169 were made using the Quikchange site-directed mutagenesis kit (Agilent Technologies). RGS2 Δ79–116 was made by inverse PCR using phosphorothioate-modified primers (Stoynova et al., 2004). The purified RGS2 peptide was made by custom peptide synthesis, and the RGS2-ala mutants were made by custom gene synthesis (Genscript Corporation). Anti-eIF2Bϵ and protein A/G PLUS-agarose beads were purchased from Santa Cruz Biotechnology, Inc. Anti-RGS2 was purchased from Genway Biotech, Inc. Protein G–agarose beads were purchased from GE Healthcare. Anti-Flag M2 monoclonal antibody and other reagents were purchased from Sigma-Aldrich.
Mouse hepatocyte isolation and culture
RGS2−/− mice were provided by J. Penninger (Institute of Molecular Biotechnology, Vienna, Austria). Experiments involving RGS2−/− mice were approved by the Council on Animal Care at the University of Western Ontario and adhered to the guidelines of the Canadian Council of Animal Care. Male mice (1–2-mo old) were anesthetized with ketamine/xylazine (100 mg/kg; 5 mg/kg), and livers were exposed and perfused through the vena cava with Ca2+-free Hanks' balanced solution at a rate of 5 ml/min for 10 min followed by serum-free Williams' medium containing 50 U/ml collagenase (type II; Worthington Biochemical Corporation), 10 mM Hepes, and 0.004 N NaOH at a rate of 5 ml/min for 15 min. The livers were removed and hepatocytes separated by Percoll gradient separation (GE Healthcare). Hepatocytes were cultured in Williams' medium supplemented with 10% FBS for 24 h before analysis of de novo protein synthesis.
Cell culture
MEF eIF2 S/S and eIF2 A/A cells were provided by R. Kaufman (University of Michigan, Ann Arbor, MI) and maintained in DME supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 1× essential amino acids, and 1× nonessential amino acids. Rat UMR-106 osteoblast and H9c2 cardiomyoblast cell lines were obtained from the American Type Culture Collection. UMR-106 and MCF-7 cells were grown in MEM α medium supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml amphotericin B. H9c2 cells were grown in DME supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml amphotericin B. Cells were grown in a humidified incubator in the presence of 5% CO2 at 37°C.
Adenovirus construction
Adenoviruses encoding GFP, RGS2, RGS4, and the RGS2 minigene were prepared according to the manufacturer's protocol (Microbix Biosystems, Inc.). Adenoviral titers were determined using the AdEasy Viral Titer kit according to the manufacturer's guidelines (Agilent Technologies).
Yeast two-hybrid assay
RGS2-binding partners were identified by screening against a mouse brain cDNA library using a yeast two-hybrid system (DupLEX-A; OriGene Technologies) according to the manufacturer's protocol.
RGS protein purification
Histidine-tagged RGS1 and RGS4 proteins were expressed in Escherichia coli BL21 (DE3) strain and purified to >95% purity by nickel affinity chromatography followed by gel filtration chromatography using a 10/30 column (Superdex 75 HR; GE Healthcare) as follows: 4 liters of bacterial culture were incubated with vigorous shaking at 37°C to an OD600 of 0.55. Expression of the RGS proteins was induced by the addition of 1 mM isopropylthio-β-d-galactoside for 3 h before harvesting the bacteria by centrifugation. Bacteria were resuspended in 60 ml buffer A (50 mM Hepes, pH 8.0, 150 mM NaCl, 20 mM β-mercaptoethanol, 0.1 mM PMSF, 10 µg/ml leupeptin, 1 µg/ml aprotinin, and 1% Triton X-100), lysozyme was added to a concentration of 0.2 mg/ml, and the suspension was mixed and incubated on ice for 30 min. The suspension was incubated for an additional 30 min on ice after the addition of 25 µg/ml DNase and 0.5 mM MgCl2. The mixture was centrifuged at 140,000 g for 30 min, and the volume of the supernatant was increased to 100 ml with buffer A supplemented with glycerol and imidazole (final concentrations of 20% and 20 mM, respectively). A 50% slurry of 1.5 ml equilibrated nickel-nitrilotriacetic acid affinity resin was added, and the mixture was incubated on a rocking platform for 1.5 h at 4°C, loaded onto a 30-ml column, washed with 30 ml of buffer A containing 0.5 M NaCl, and washed with 30 ml of buffer A without Triton X-100. Proteins were eluted with buffer A containing 200 mM imidazole. These protein samples were loaded and run through a 10/30 column (Superdex 75 HR) equilibrated with buffer B (50 mM Hepes, pH 8.0, 150 mM NaCl, 1 mM DTT, and 0.1 mM PMSF), and the peak fractions were collected and stored at −80°C.
Histidine-tagged RGS2 and all its derivative mutants were purified from bacterial inclusion bodies as follows: 2 liters BL21 (DE3) bacterial culture was incubated with vigorous shaking at 37°C until mid-log phase. Induction was commenced by the addition of 1 mM isopropylthio-β-d-galactoside for 3–4 h at 37°C. The bacteria were harvested by centrifugation, and pellets were stored at −80°C. Cells were thawed and resuspended in 30 ml of IB (inclusion body) buffer (20 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.1 mM PMSF, 10 µg/ml leupeptin, 1 µg/ml aprotinin, and 1% Triton X-100), and 0.2 mg/ml lysozyme was added. The suspension was mixed and incubated at 30°C for 15 min followed by sonication on ice. Cell lysates were centrifuged at 15,000 g for 30 min at 4°C, and the pellet was washed by resuspension and centrifugation once with IB buffer and once with 0.1 M Tris-HCl, pH 8.0. The protein-refolding process was accomplished by extracting inclusion bodies with extraction buffer (10 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 6 M guanidine hydrochloride) followed by dialysis against buffer A (50 mM Hepes, pH 7.5, 500 mM NaCl, 20 mM β-mercaptoethanol, 10% glycerol, 0.5 mM PMSF, and 2 M urea) and buffer B (50 mM Hepes, pH 7.5, 500 mM NaCl, 20 mM β-mercaptoethanol, 10% glycerol, 0.5 mM PMSF, and 500 mM urea). The refolded protein was purified by nickel chromatography followed by size-exclusion chromatography as described in the previous paragraph for RGS1 and RGS4 except that the concentration of NaCl was 0.5 M throughout the imidazole elution step and reduced to 0.3 M for the final gel filtration step. For RGS2-ala1 and RGS2-ala2, the proteins were purified by nickel chromatography and dialyzed with storage buffer (50 mM Hepes, pH 7.5, 500 mM NaCl, 20 mM β-mercaptoethanol, 0.1 mM PMSF, 1 µg/ml aprotinin, 10 µg/ml leupeptin, and 0.5 mM urea) to remove imidazole. Protein samples were placed in aliquots and stored at −80°C.
GTP hydrolysis assay
Sf9 insect cells at a density of 2 × 106 cells/ml were infected with baculoviruses encoding Gβ1, Gγ2, and an M1 muscarinic receptor–Gα11 fusion protein. At 48 h after infection, cells were centrifuged at 228 g for 5 min, resuspended in PBS, and centrifuged again. The resulting pellet was resuspended in one third of the original volume of lysis buffer (20 mM Tris, pH 8.0, 0.1 mM PMSF, 10 µg/ml leupeptin, and 1 µg/ml aprotinin) and incubated on ice for 15 min. The cells were lysed using a homogenizer (Polytron; Brinkmann Instruments) and centrifuged at 500 g for 10 min. The supernatant was retained and centrifuged for 30 min at 48,000 g. The supernatant from this centrifugation was discarded, and the pellets were resuspended in 0.01 vol of lysis buffer and stored at −80°C.
Membranes were assayed for carbachol-stimulated GTP hydrolysis for 5 min at 30°C in the absence and presence of the indicated RGS proteins. The reaction buffer contained 106 cpm/assay γ-[32P]GTP, 20 mM Hepes, pH 7.5, 1 mM EDTA, 1 mM DTT, 200 nM GTP, 0.5 mM ATP, 0.1 mM PMSF, 1 µg/ml aprotinin, 10 µg/ml leupeptin, 0.1 mM ascorbic acid, 300 mM NaCl, and 2 mM MgCl2. Nonspecific GTPase activity was defined as that in the presence of membranes plus the inverse agonist, 10 µM tropicamide, and these values were subtracted to yield the specific agonist- and receptor-dependent signal. GTP hydrolysis reactions were terminated by the addition of 5% Norit in 50 mM NaH2PO4, pH 3.0. The reaction mixture was centrifuged at 2,000 g, and 32Pi was recovered from the supernatant. Radioactivity was measured on a liquid scintillation counter (Packard Tri-Carb 2900TR; PerkinElmer).
In vitro translation assay
Translation was measured as the synthesis of luciferase protein from luciferase mRNA using an in vitro translation kit (Applied Biosystems) according to the manufacturer's protocol. In brief, rabbit reticulocyte lysates were combined with 20 µM amino acid mixture, 0.5 µg luciferase RNA (Promega), 1× low salt translation mix, and RGS proteins where indicated. The final concentration of NaCl in the assay was equalized to 300 mM by dilution. The mixture was incubated at 30°C for 45 min, and luminescence was detected with luciferase assay substrate (Promega) using a microplate reader (LMax II; MDS Analytical Technologies).
Cellular protein synthesis assay
Cells were seeded in 12-well cluster plates at a density of 500,000 cells/well. 24 h later, cells were infected with the indicated virus. At 48 h after infection, the growth medium was removed and replaced with reduced-serum medium (Opti-MEM I; Invitrogen) supplemented with 0.5 µCi/ml [3H]leucine for 4 h. Cells were washed with PBS and incubated on ice with ice-cold lysis buffer (20 mM Hepes, pH 7.5, 50 mM KCl, 0.1 mM PMSF, 1 µg/ml aprotinin, 10 µg/ml leupeptin, 0.1 mM EDTA, 10% [vol/vol] glycerol, and 1% Triton X-100) for 15 min. Protein concentrations were measured using a BSA protein assay (Bio-Rad Laboratories). An equal volume of the cell lysate was combined with a 25% (wt/vol) solution of trichloroacetic acid to precipitate the protein, subjected to a vigorous vortex, and incubated on ice for 30 min. The mixture was filtered through a filtration apparatus (Millipore) onto glass microfiber disks (GF/C; GE Healthcare) and washed three times each with 5% TCA followed by 100% ethanol. Disks were air dried overnight, and their radioactivity was measured using a liquid scintillation counter (Packard Tri-Carb 2900TR; PerkinElmer).
eIF2B GEF Assay
The ability of eIF2B to promote the dissociation of GDP from eIF2 was measured as described previously (Kimball et al., 1991). In brief, eIF2 and eIF2B were purified from rat liver (Kimball et al., 1987), and the rate of exchange of [3H]GDP bound to eIF2 for free, nonradioactively labeled GDP was measured in the absence and presence of the indicated purified RGS2 proteins. The activity of eIF2B was calculated as the slope of the nearest fit line with dpm as the dependent variable and time as the independent variable.
Coimmunoprecipitation
For in vitro coimmunoprecipitations, 106 cells/ml Sf9 insect cells were infected with recombinant baculovirus encoding Flag-tagged eIF2Bϵ subunit alone or coinfected with the three recombinant baculovirus stocks encoding all five eIF2B subunits (Fabian et al., 1997). At 72 h after infection, cells were washed once with ice-cold PBS and lysed with immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and 10% glycerol). Cellular debris was removed by centrifugation at 14,000 g for 10 min at 4°C, and these lysates were mixed with 10 µg of RGS protein and subsequently precleared by mixing with protein G beads for 1 h at 4°C and then centrifuged. The precleared lysate mixture was incubated with 2 µg anti-Flag M2 monoclonal antibody for 3h at 4°C. The protein G beads were washed three times with immunoprecipitation buffer and were resuspended in 2× Laemmli sample buffer.
For coimmunoprecipitation of endogenous proteins, confluent UMR-106 cells in a T75 tissue culture flask were incubated for 3 h at 37°C with vehicle control or 100 µM forskolin to up-regulate RGS2 expression essentially as described previously (Roy et al., 2006b). Cells were washed twice with ice-cold PBS and lysed by incubating with ice-cold hypotonic buffer (20 mM Hepes, pH 7.5, 50 mM KCl, 0.1 mM PMSF, 1 µg/ml aprotinin, 10 µg/ml leupeptin, 0.1 mM EDTA, 10% [vol/vol] glycerol, and 1% Triton X-100) for 30 min. Cellular lysates were centrifuged at 10,000 g, and the supernatant was removed and precleared by rotating with 1 µg mouse IgG and 20 µl resuspended protein A/G PLUS-agarose for 30 min at 4°C. The precleared cellular extract (1 mg total cellular protein) was rotated with 2 µg anti-eIF2Bϵ for 1 h at 4°C, and 25 µl protein A/G PLUS-agarose was subsequently added for an overnight incubation. Immunocomplexes were washed five times with PBS and resuspended in 2× Laemmli sample buffer.
Immunoblot analysis
Samples were placed in boiling water for 5 min and run on a 12% SDS polyacrylamide gel, transferred to a polyvinylidene fluoride membrane, and probed with the indicated antibodies according to the manufacturer's recommendations. Blots were visualized by chemiluminescence substrate (LumiGLO Reserve; Kirkegaard & Perry Laboratories, Inc.) using an imaging system (Fluorchem 8000; Alpha Innotech Corporation). Images were processed using Photoshop (CS2; Adobe) to adjust for brightness and contrast only.
Acknowledgments
We are grateful to our colleagues who provided experimental materials, in particular Dr. Qingshi Zhao who made the RGS1 polyhistidine construct, Dr. Qingming Ding for generating the adenovirus, and Dr. Randall Kaufman, who provided the eIF2S/S and eIF2A/A MEF cell lines. We also thank members of the Chidiac laboratory and Dr. Stanley Dunn for helpful discussions.
P. Chidiac holds a Career Investigator Award from the Heart and Stroke Foundation of Ontario. This work was supported by operating grants from the Heart and Stroke Foundation of Ontario, the Canadian Institutes of Health Research (P. Chidiac and R. Gros), and the U.S. National Institutes of Health (grant DK-15658 to S.R. Kimball).
Footnotes
Abbreviations used in this paper:
- GAP
- GTPase-accelerating protein
- GEF
- guanine nucleotide exchange factor
- MEF
- mouse embryonic fibroblast
- RGS
- regulator of G protein signaling
References
- Bernstein L.S., Ramineni S., Hague C., Cladman W., Chidiac P., Levey A.I., Hepler J.R. 2004. RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling.J. Biol. Chem. 279:21248–21256 [DOI] [PubMed] [Google Scholar]
- Boesen T., Mohammad S.S., Pavitt G.D., Andersen G.R. 2004. Structure of the catalytic fragment of translation initiation factor 2B and identification of a critically important catalytic residue.J. Biol. Chem. 279:10584–10592 [DOI] [PubMed] [Google Scholar]
- Deng J., Harding H.P., Raught B., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N. 2002. Activation of GCN2 in UV-irradiated cells inhibits translation.Curr. Biol. 12:1279–1286 [DOI] [PubMed] [Google Scholar]
- Dever T.E. 2002. Gene-specific regulation by general translation factors.Cell. 108:545–556 [DOI] [PubMed] [Google Scholar]
- Fabian J.R., Kimball S.R., Heinzinger N.K., Jefferson L.S. 1997. Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells.J. Biol. Chem. 272:12359–12365 [DOI] [PubMed] [Google Scholar]
- Gebauer F., Hentze M.W. 2004. Molecular mechanisms of translational control.Nat. Rev. Mol. Cell Biol. 5:827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Anton A., Salim S., Blumer K.J., Dessauer C.W., Heximer S.P. 2008. Alternative translation initiation of human regulators of G-protein signaling-2 yields a set of functionally distinct proteins.Mol. Pharmacol. 73:1–11 [DOI] [PubMed] [Google Scholar]
- Gutiérrez P., Osborne M.J., Siddiqui N., Trempe J.F., Arrowsmith C., Gehring K. 2004. Structure of the archaeal translation initiation factor aIF2 beta from Methanobacterium thermoautotrophicum: implications for translation initiation.Protein Sci. 13:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A.P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S.H., Chen J.J. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency.EMBO J. 20:6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt S.E., Tomita H., Katus H.A., Sadoshima J. 2004. Phosphorylation of eukaryotic translation initiation factor 2Bepsilon by glycogen synthase kinase-3beta regulates beta-adrenergic cardiac myocyte hypertrophy.Circ. Res. 94:926–935 [DOI] [PubMed] [Google Scholar]
- Heo K., Ha S.H., Chae Y.C., Lee S., Oh Y.S., Kim Y.H., Kim S.H., Kim J.H., Mizoguchi A., Itoh T.J., et al. 2006. RGS2 promotes formation of neurites by stimulating microtubule polymerization.Cell. Signal. 18:2182–2192 [DOI] [PubMed] [Google Scholar]
- Ingi T., Krumins A.M., Chidiac P., Brothers G.M., Chung S., Snow B.E., Barnes C.A., Lanahan A.A., Siderovski D.P., Ross E.M., et al. 1998. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity.J. Neurosci. 18:7178–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S.R. 1999. Eukaryotic initiation factor eIF2.Int. J. Biochem. Cell Biol. 31:25–29 [DOI] [PubMed] [Google Scholar]
- Kimball S.R., Everson W.V., Myers L.M., Jefferson L.S. 1987. Purification and characterization of eukaryotic initiation factor 2 and a guanine nucleotide exchange factor from rat liver.J. Biol. Chem. 262:2220–2227 [PubMed] [Google Scholar]
- Kimball S.R., Antonetti D.A., Brawley R.M., Jefferson L.S. 1991. Mechanism of inhibition of peptide chain initiation by amino acid deprivation in perfused rat liver. Regulation involving inhibition of eukaryotic initiation factor 2 alpha phosphatase activity.J. Biol. Chem. 266:1969–1976 [PubMed] [Google Scholar]
- Kimball S.R., Heinzinger N.K., Horetsky R.L., Jefferson L.S. 1998. Identification of interprotein interactions between the subunits of eukaryotic initiation factors eIF2 and eIF2B.J. Biol. Chem. 273:3039–3044 [DOI] [PubMed] [Google Scholar]
- Klein U., Ramirez M.T., Kobilka B.K., von Zastrow M. 1997. A novel interaction between adrenergic receptors and the alpha-subunit of eukaryotic initiation factor 2B.J. Biol. Chem. 272:19099–19102 [DOI] [PubMed] [Google Scholar]
- Lu L., Han A.P., Chen J.J. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses.Mol. Cell. Biol. 21:7971–7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchy D.B., Knudsen C.R., Clark B.F., Kahn R.A., Hall R.A., Levey A.I. 2002. Novel interaction between the M4 muscarinic acetylcholine receptor and elongation factor 1A2.J. Biol. Chem. 277:29268–29274 [DOI] [PubMed] [Google Scholar]
- McClatchy D.B., Fang G., Levey A.I. 2006. Elongation factor 1A family regulates the recycling of the M4 muscarinic acetylcholine receptor.Neurochem. Res. 31:975–988 [DOI] [PubMed] [Google Scholar]
- Mohammad-Qureshi S.S., Haddad R., Hemingway E.J., Richardson J.P., Pavitt G.D. 2007. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2beta and -2gamma mediate guanine nucleotide exchange.Mol. Cell. Biol. 27:5225–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezeau L., Richman J.G., Edwards S.W., Limbird L.E. 1999. The zeta isoform of 14-3-3 proteins interacts with the third intracellular loop of different alpha2-adrenergic receptor subtypes.J. Biol. Chem. 274:13462–13469 [DOI] [PubMed] [Google Scholar]
- Proud C.G. 2005. eIF2 and the control of cell physiology.Semin. Cell Dev. Biol. 16:3–12 [DOI] [PubMed] [Google Scholar]
- Proud C.G. 2007. Signalling to translation: how signal transduction pathways control the protein synthetic machinery.Biochem. J. 403:217–234 [DOI] [PubMed] [Google Scholar]
- Ron D. 2002. Translational control in the endoplasmic reticulum stress response.J. Clin. Invest. 110:1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E.M., Wilkie T.M. 2000. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins.Annu. Rev. Biochem. 69:795–827 [DOI] [PubMed] [Google Scholar]
- Roy A.A., Baragli A., Bernstein L.S., Hepler J.R., Hébert T.E., Chidiac P. 2006a. RGS2 interacts with Gs and adenylyl cyclase in living cells.Cell. Signal. 18:336–348 [DOI] [PubMed] [Google Scholar]
- Roy A.A., Nunn C., Ming H., Zou M.X., Penninger J., Kirshenbaum L.A., Dixon S.J., Chidiac P. 2006b. Up-regulation of endogenous RGS2 mediates cross-desensitization between Gs and Gq signaling in osteoblasts.J. Biol. Chem. 281:32684–32693 [DOI] [PubMed] [Google Scholar]
- Salim S., Sinnarajah S., Kehrl J.H., Dessauer C.W. 2003. Identification of RGS2 and type V adenylyl cyclase interaction sites.J. Biol. Chem. 278:15842–15849 [DOI] [PubMed] [Google Scholar]
- Santos de Araujo R.M., Oba Y., Moriyama K. 2007. Role of regulator of G-protein signaling 2 (RGS2) in periodontal ligament cells under mechanical stress.Cell Biochem. Funct. 25:753–758 [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R.J. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis.Mol. Cell. 7:1165–1176 [DOI] [PubMed] [Google Scholar]
- Schoeber J.P., Topala C.N., Wang X., Diepens R.J., Lambers T.T., Hoenderop J.G., Bindels R.J. 2006. RGS2 inhibits the epithelial Ca2+ channel TRPV6.J. Biol. Chem. 281:29669–29674 [DOI] [PubMed] [Google Scholar]
- Shan D., Wang H., Su Y., Jing Y., Wong T.M. 2007. kappa-opioid receptor stimulation inhibits cardiac hypertrophy induced by beta1-adrenoceptor stimulation in the rat.Eur. J. Pharmacol. 555:100–105 [DOI] [PubMed] [Google Scholar]
- Siderovski D.P., Heximer S.P., Forsdyke D.R. 1994. A human gene encoding a putative basic helix-loop-helix phosphoprotein whose mRNA increases rapidly in cycloheximide-treated blood mononuclear cells.DNA Cell Biol. 13:125–147 [DOI] [PubMed] [Google Scholar]
- Singh C.R., Lee B., Udagawa T., Mohammad-Qureshi S.S., Yamamoto Y., Pavitt G.D., Asano K. 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation.EMBO J. 25:4537–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnarajah S., Dessauer C.W., Srikumar D., Chen J., Yuen J., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H. 2001. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III.Nature. 409:1051–1055 [DOI] [PubMed] [Google Scholar]
- Song L., Jope R.S. 2006. Cellular stress increases RGS2 mRNA and decreases RGS4 mRNA levels in SH-SY5Y cells.Neurosci. Lett. 402:205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan M., Willard F.S., Kimple A.J., Turnbull A.P., Ball L.J., Schoch G.A., Gileadi C., Fedorov O.Y., Dowler E.F., Higman V.A., et al. 2008. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.Proc. Natl. Acad. Sci. USA. 105:6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoynova L., Solórzano R., Collins E.D. 2004. Generation of large deletion mutants from plasmid DNA.Biotechniques. 36:402–404: 406 [DOI] [PubMed] [Google Scholar]
- Wek R.C., Jiang H.Y., Anthony T.G. 2006. Coping with stress: eIF2 kinases and translational control.Biochem. Soc. Trans. 34:7–11 [DOI] [PubMed] [Google Scholar]
- Welsh G.I., Miller C.M., Loughlin A.J., Price N.T., Proud C.G. 1998. Regulation of eukaryotic initiation factor eIF2B: glycogen synthase kinase-3 phosphorylates a conserved serine which undergoes dephosphorylation in response to insulin.FEBS Lett. 421:125–130 [DOI] [PubMed] [Google Scholar]
- Zhang W., Anger T., Su J., Hao J., Xu X., Zhu M., Gach A., Cui L., Liao R., Mende U. 2006. Selective loss of fine tuning of Gq/11 signaling by RGS2 protein exacerbates cardiomyocyte hypertrophy.J. Biol. Chem. 281:5811–5820 [DOI] [PubMed] [Google Scholar]
- Zmijewski J.W., Song L., Harkins L., Cobbs C.S., Jope R.S. 2001. Oxidative stress and heat shock stimulate RGS2 expression in 1321N1 astrocytoma cells.Arch. Biochem. Biophys. 392:192–196 [DOI] [PubMed] [Google Scholar]
- Zou M.X., Roy A.A., Zhao Q., Kirshenbaum L.A., Karmazyn M., Chidiac P. 2006. RGS2 is upregulated by and attenuates the hypertrophic effect of alpha1-adrenergic activation in cultured ventricular myocytes.Cell. Signal. 18:1655–1663 [DOI] [PubMed] [Google Scholar]