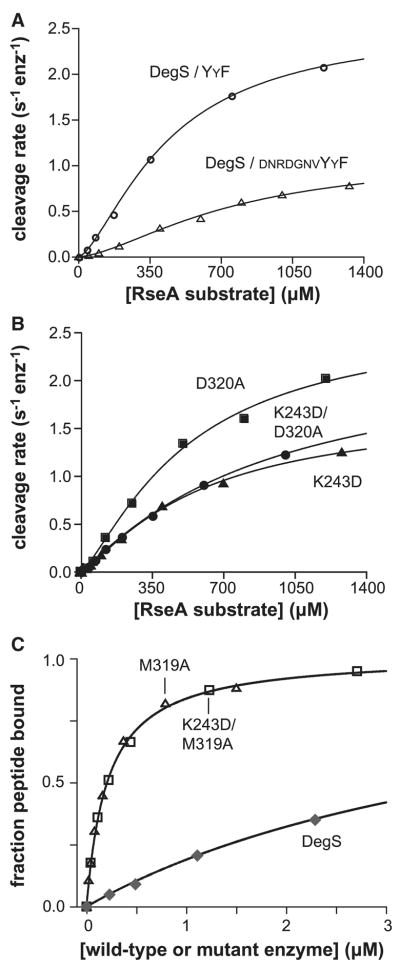
Figure 3.
Substrate binding and control of activity.
(A) Substrate dependence of the steady-state rate of cleavage by wild-type DegS (0.5 μM trimer) with YYF peptide (300 μM). For comparison, rates in the presence of saturating DNRDGNVYYF peptide (Sohn et al., 2007) are also shown. (B) Substrate dependence of the cleavage by the K243D, D320A and K243D/D320A mutants of DegS (0.3 μM trimer) with DNRDGNVYYF peptide (30 μM). (C) Binding of DegS, the M319A mutant, and the K243D/M319A mutant to the OMP peptide fluoresceine-β–alanine-KKDNRDGNYYF (20 nM) were monitored by changes in fluorescence anisotropy. The lines are fits to a quadratic form of a hyperbolic binding isotherm. The data for wild-type DegS are from Sohn et al. (2007). In panels A & B, the lines are fits to the Hill form of the Michaelis-Menten equation: velocity = Vmax/(1+(KM/[substrate])n).