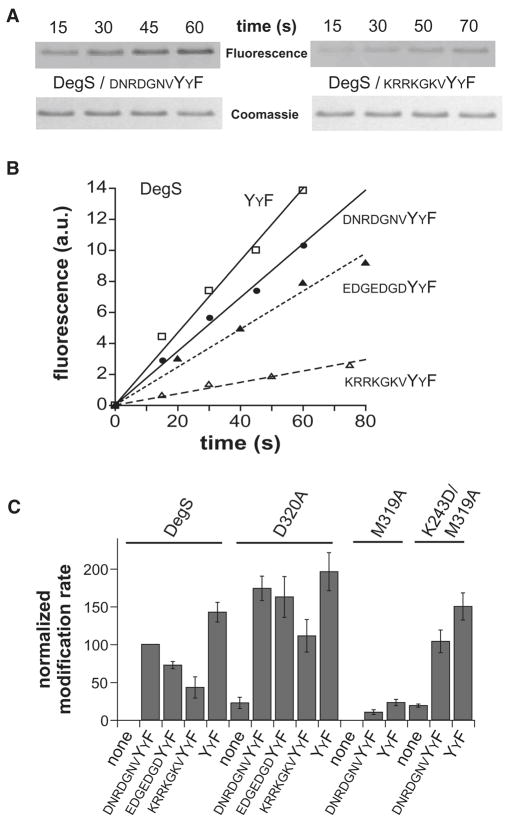
Figure 4.
Peptide dependence of active-site modification.
(A) SDS-PAGE of wild-type DegS after reaction with rhodamine-FP in the presence of two OMP peptides. Fluorescence is shown in the upper strips, and Coomassie-blue staining in the lower strips. (B) Rhodamine-FP (20 μM) modification of wild-type DegS (0.9 μM trimer) in the presence of saturating concentrations of four OMP-like peptides. The lines are linear fits (R ≥ 0.975). (C) Rates of rhodamine-FP modification of wild-type DegS and the D320A, M319A, and K243D/M319A mutants without peptide or with saturating OMP peptides. Rates were normalized to an arbitrary value of 100 for wild-type DegS with saturating DNRDGNVYYF peptide. The error bars represent standard deviations based on three or more experiments.