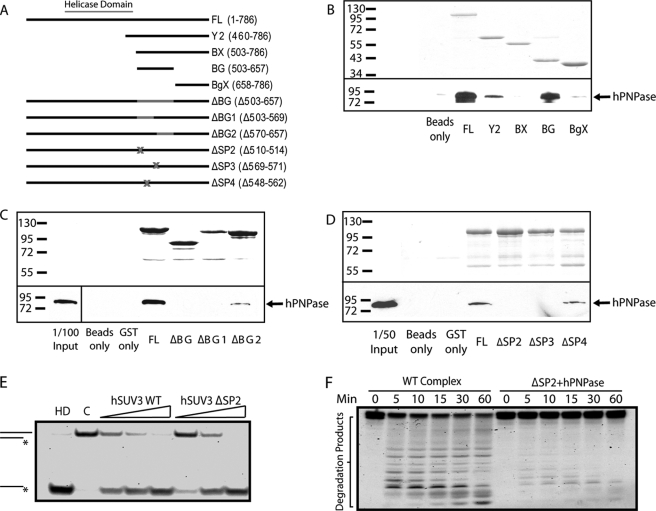
FIGURE 3.
Complex formation is critical for hSUV3 to interact with PNPase to degrade structured RNA. A, list of GST-hSUV3 constructs used in the in vitro binding experiments. B–D, the top panels show the Coomassie Blue staining of the input hSUV3 recombinant proteins. The bottom panels show the amount of hPNPase pulled down by the hSUV3 proteins and detected by Western blotting using monoclonal anti-hSUV3 antibody. E, increasing amounts of wild type hSUV3 and ΔSP2 (30, 60, and 120 nm) were incubated with the 3′-OH helicase substrate for 30 min at 37 °C in the presence of 5 mm ATP. Lane HD, heat denatured. Lane C, control. F, time course 3′-OH dsRNA substrate degradation activities of the hSUV3-hPNPase complex (120 nm) and a mixture containing hSUV3 ΔSP2 and hPNPase of equivalent molar amounts in the presence 3 mm ATP and 5 mm phosphate.