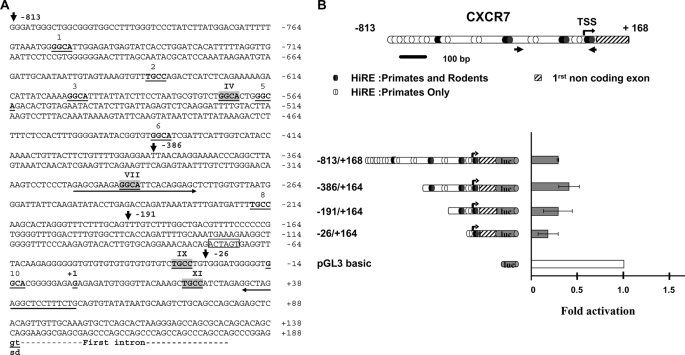
FIGURE 4.
CXCR7 is down-regulated by HIC1. A, nucleotide sequence of the 5′ region of the human CXCR7 gene. The transcription start site as well as the first noncoding exon are described in GenBankTM under accession number NM_020311; gi 114155149. The conserved gt dinucleotides of the splice donor site (sd) are indicated in boldface letters and underlined. Upstream genomic sequences were extracted from the human genome sequence (NC_000002). Potential HiREs are indicated with the 4-bp core consensus 5′-GGCA-3′ (reverse complement strand TGCC) shown in boldface and underlined (5). These sites are numbered from 1 to 11. The sites indicated by roman numerals and highlighted in light gray boxes are those conserved in the primate (human and chimpanzee) and rodent (mice and rat) genomes (see also supplemental Fig. 3 for a CLUSTAL alignment). The 5′ boundaries of the various deletions in the CXCR7 promoter obtained by PCR are indicated by arrows with the numbering relative to the +1 position of the transcription initiation site. A unique SpeI site (position −75 to −70) used to obtain the −863/+168 ΔXI construct is boxed. Primers used to amplify the CXCR7 promoter fragment in the ChIP experiment presented in Figs. 6 and 7 are indicated by an arrow. B, from top to bottom, a schematic drawing of the 5′ promoter region and the first noncoding exon of CXCR7 is shown. Numbering is relative to the transcription start site (TSS) (bent arrow, position +1). The potential HiREs (5) are shown as black ovals for those found in the primate and rodent genomes or as white ovals for those found only in the primate genomes. The small arrows indicate the position of the primers used to amplify the relevant region of CXCR7 in the ChIP and ChIP upon ChIP experiments. The left panel shows a schematic drawing of various fragments of the human CXCR7 first noncoding exon 5′ sequences subcloned in the luciferase reporter plasmid pGL3 basic. Reporter constructs were transfected in triplicate into U2OS cells and assayed for luciferase activity. Luciferase (luc) activity was normalized for transfection efficiency using a co-transfected β-galactosidase reporter. Repression of transcription of each construct by HIC1 was calculated by first dividing luciferase activity in the absence of HIC1 by the activity in the presence of HIC1 (normalized for transfection efficiency using a co-transfected β-galactosidase reporter). The value obtained for each construct was then divided by the repressive effect elicited by HIC1 on the empty pGL3 basic vector to obtain the final fold of activation. Results, expressed relative to a value of 1.0 for cells transfected with the pGL3 empty vector, are expressed as the mean of three different experiments, and error bars represent standard deviations.