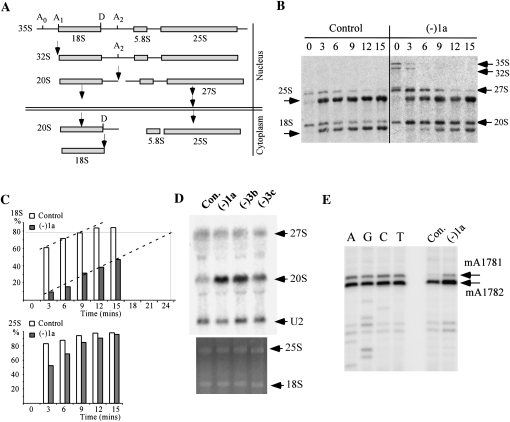
FIGURE 6.
Disrupting the hypermodification strongly delays hydrolytic processing of pre-rRNA. (A) Schematic representation of the yeast pre-rRNA processing pathway. Cleavage sites involved in formation of 18S rRNA are shown. The processing steps for 27S pre-rRNA are depicted in a simplified form. (B) Radiolabeling pulse-chase analysis shows that pre-rRNA processing is delayed in cells lacking the hypermodified Ψ. Pre-rRNA was labeled in vivo using [3H]methionine, and processing products were analyzed on 1.2% agarose gels and visualized with a PhosphorImager. The chase times (minutes) are indicated above the lanes. (C) Percentages of converted mature 18S rRNA (upper panel) and 25S rRNA (lower panel) at each time point. Signal strength was measured from the results in panel B using ImageJ, and the calculated values plotted. (D) Loss of the hypermodified Ψ causes substantial accumulation of 20S pre-rRNA. Steady state levels of rRNA species were examined by Northern hybridization, with oligonucleotides specific to 20S and 27S pre-rRNAs. U2 snRNA was used as a control for sample loading. (E) The 20S pre-rRNA lacking the hypermodified Ψ is exported to the cytoplasm. The presence of the cytoplasmic dimethylations in the 20S pre-rRNA was detected by primer extension analysis. The reaction products were separated on an 8% polyacrylamide gel, next to a sequencing ladder created with the same oligonucleotide.