Figure 2. Phylogenetic analysis of Xenopus laevis class Ib.
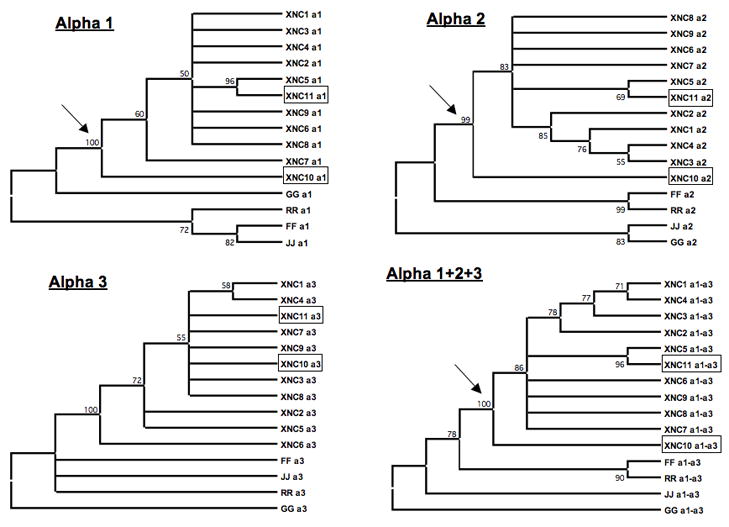
Sequences were aligned with ClustalX and phylogenetic analyses were performed using Molecular Evolutionary Genetics Analysis (MEGA; version 3.1). Phylogenetic trees were constructed using the neighbor-joining method with pairwise deletion of gaps using MEGA3.1 software (Kumar et al, 2001). Support for each node was assessed using 1000 bootstrap replicates. Accession numbers: XNC1 [GenBank:M58019], 2 [GenBank:L20725], 3 [GenBank:L20726], 4 [GenBank:L20727], 5 [GenBank:L20728], 6 [GenBank:L20729], 7 [GenBank:L20730], 8 [GenBank:L20731], 9 [GenBank:L20732], 10 [GenBank:bankit1167220], and 11 [GenBank:bankit1167224]; class Ia G [GenBank:AF185579], R [GenBank:AF185582], F [GenBank:AAF03402], and J [GenBank:AF185586].
