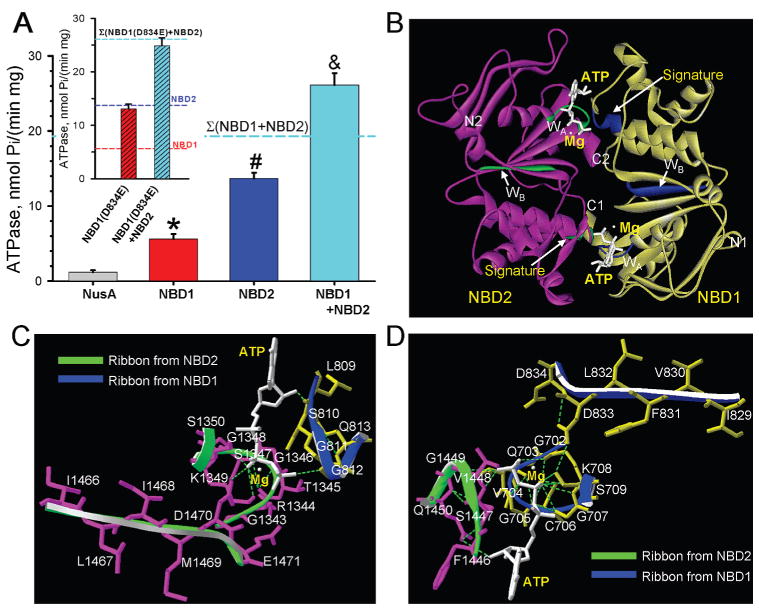
Figure 4.
NBD1/NBD2 assembly promotes intrinsic ATPase activity. (A) The ATPase activity of NBD1 or NBD2 was significantly higher than background ascribed to the NusA tag. Upon NBD1/NBD2 interaction, ATPase activity significantly increased, exceeding the sum of individual NBD1 + NBD2 catalytic activities. *, #, and & indicate significantly greater (P < 0.05) than NusA, than NusA or NBD1, and NusA, NBD1, or NBD2, respectively. Inset: The D834E mutation increased NBD1 ATPase activity approximating wildtype NBD2 catalysis. NBD1 (D834E) and NBD2, free to interact, produced a total ATPase that did not exceed the arithmetic sum of individual catalytic activities. (B) Homology structural model of the NBD1/NBD2 heterodimer, developed in the “head to tail” orientation, indicated that Walker A (WA) and Walker B (WB) motifs along with a signature motif delineate nucleotide docking sites. NBD1 in yellow, NBD2 in purple, and the ATPase cofactor Mg2+ in white. Hydrogen bonds shown as dotted lines. N- and C-termini of individual NBDs are labeled. Close-up view of ATP-binding sites at the “head” (C) and “tail” (D) of the heterodimer.