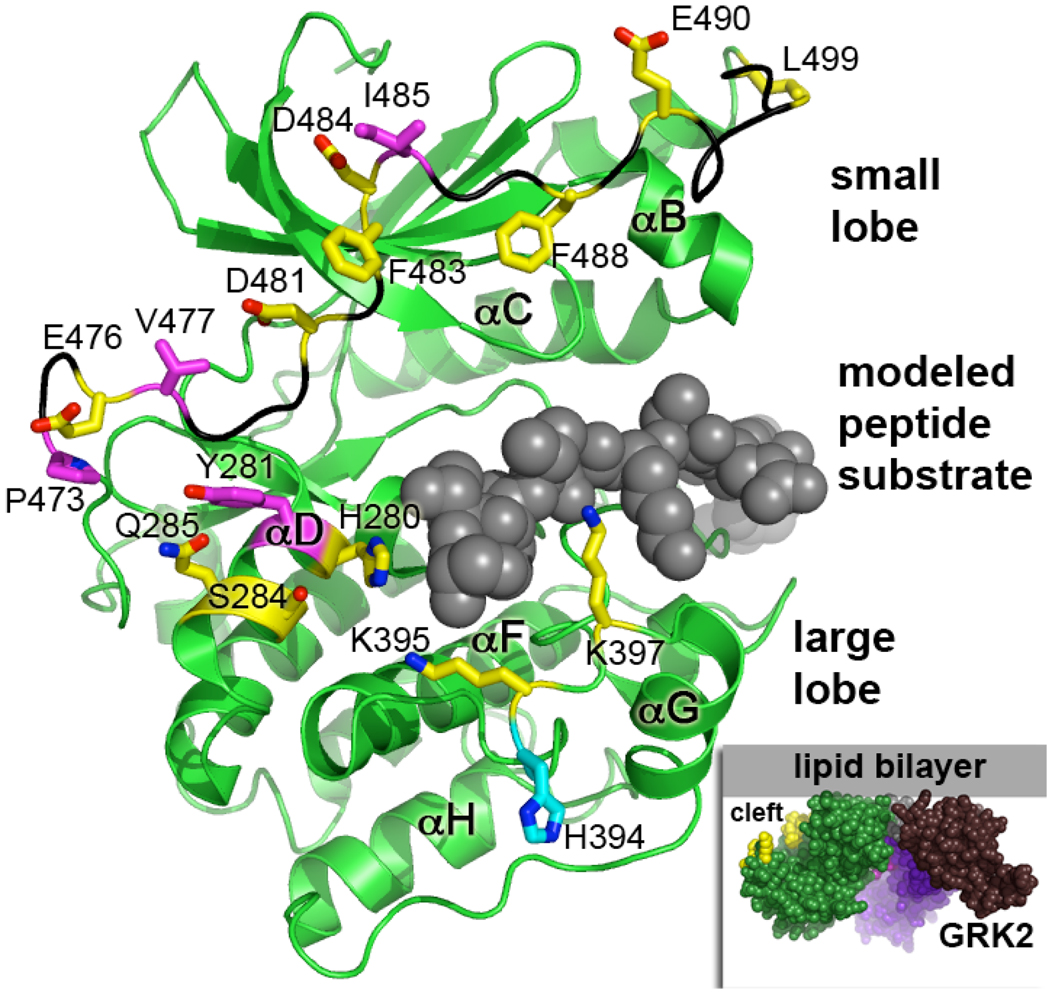
Figure 2. Kinase Domain Residues Targeted for Mutagenesis.
The GRK2 kinase domain was modeled in a closed conformation based on an activated structure of PKA (PDB 1L3R). Two regions of the kinase domain were targeted for mutagenesis: the large lobe and the C-tail. To indicate the expected position of the phosphoacceptor binding site, the PKA inhibitor PKI (space-filling atoms in gray) was mapped onto the GRK2 structure. Nearby residues that could interact with other regions of the receptor (His280, Tyr281, Gln285, Ser284, His394, Lys395, and Lys397) were substituted with alanine. The AST of GRK2 has also been implicated in receptor interaction (20) but residues Glu476-Leu499 have been unstructured in all crystals of GRK2 reported thus far. However, a nearly fully ordered AST loop (black backbone) was observed in GRK1 (8). This structure was mapped onto GRK2 to provide an estimate of the position of amino acids in the AST. Pro473, Glu476, Val477, Asp481, Phe483, Asp484, Ile485, Phe488, Glu490, Gly495, and Leu499 in the AST were selected for site-directed mutagenesis. Substituted residues that showed diminished capacity to phosphorylate receptor and at least some of the non-receptor substrates have carbon atoms colored magenta, whereas those that did not have significant effects on rhodopsin phosphorylation have carbons colored yellow. (Inset) A space-filling model of GRK2 at the plasma membrane using the Fig. 1 coloring scheme (RH domain is purple and magenta, and the PH domain is colored brown). We speculated that the cleft between the lipid bilayer and the kinase large lobe could serve to accommodate the intracellular loops and carboxyl tail of a GPCR. Mutated residues in the kinase domain are shown as yellow spheres.