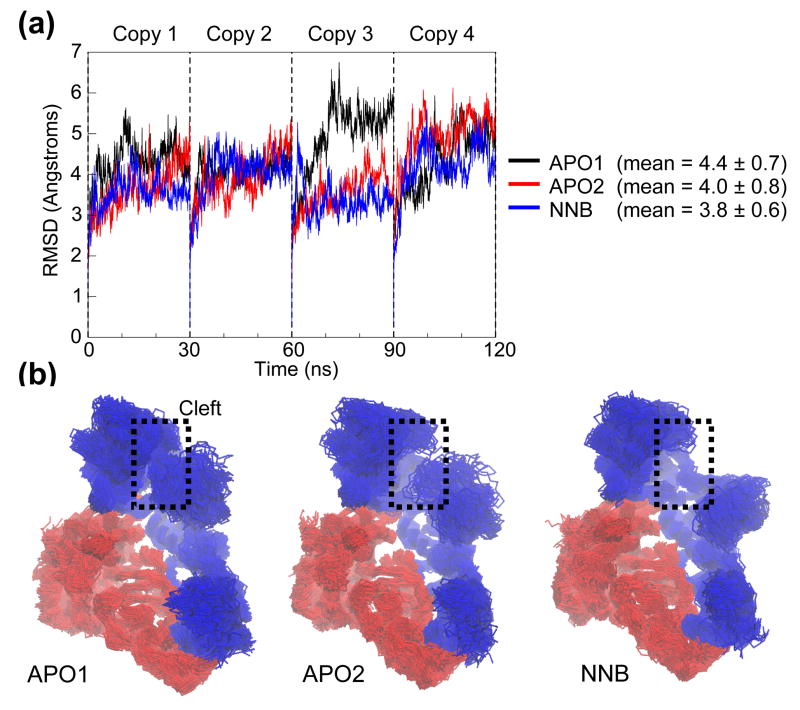
Figure 3.
Conformational drift. (a) RMSD of all Cα atoms from the starting structure, as a function of time. Each 30 ns simulation copy is concatenated into a single 120 ns trajectory for analysis. (b) Superposition of 100 RT snapshots (taken at 1.2 ns intervals) from each simulation system. Protein is shown in Cα trace representation, with p66 and p51 subunits colored blue and red, respectively. Attention is drawn to variations in the dynamics of the nucleic acid binding cleft.