Abstract
Purpose
Several reports have suggested the rat Vcsa1 gene is down-regulated in models of erectile dysfunction (ED). Vcsa’s protein product, sialorphin, is an endogenous neutral endopeptidase (NEP), and its down-regulation could result in prolonged activation of G-protein activated signaling pathways by their peptide agonists. We investigated if down- regulation of Vcsa1 could result in adaptive change in the expression of G-protein coupled receptors (GPCR).
Materials and Methods
Gene expression in cultured rat corporal smooth muscle cells (CSM) following treatment with siRNA directed against Vcsa1 or the NEP gene was analyzed using microarray and quantitative RT-PCR. In rats Vcsa1 is one of the most down-regulated genes following bilateral transection of the cavernosal nerves. Using that animal model, we also investigated whether the down-regulation of Vcsa1 is accompanied by similar changes in gene expression observed in the CSM cells where Vcsa1 was knocked-down in vitro.
Results
Microarray analysis and quantitative RT-PCR demonstrated that CSM cells treated in vitro with siRNA against Vcsa1 resulted in up-regulation of GPCR as a functional group. In contrast, treatment of CSM cells that lowered NEP activity resulted in decreases in GPCR expression. These results suggest that the peptide product of Vcsa1, sialorphin, can effect GPCR expression by acting on NEP. In animals with bilaterally transected cavernous nerves the reduced expression of Vcsa1 is accompanied by increased GPCR expression in cavernosal tissue.
Conclusions
These experiments suggest that the mechanism by which Vcsa1 modulates erectile function is partly mediated through changes in GPCR expression.
Keywords: erectile dysfunction, G-protein coupled receptors, neutral endopeptidase inhibitor, sialorphin, Vcsa1
1. Introduction
Recent reports have demonstrated that the Vcsa1 gene is one of the most down-regulated genes in the corporal tissue of several rat models of erectile dysfunction (ED) 1, 2. A human homologue of Vcsa1, hSMR3A, is also down-regulated in patients with ED 3. Vcsa1, which encodes a pentapeptide called sialorphin, belongs to a family of genes encoding peptides which act as endogenous neutral endopeptidase (NEP) inhibitors 4, 5. The Vcsa1 gene is expressed in relatively few organs, which include the submandibular and prostate glands and the corpora cavernosa, while the sialorphin peptide, is found in the circulatory system and saliva 2, 6. Sialorphin has been demonstrated to be involved in several physiological processes, such as pain perception, antidepressant effects, sexual behavior and erectile function in rats 2, 7, 8.
The diverse physiological roles of sialorphin may be a result of the inhibition of the almost ubiquitous tissue expression of its target enzyme, NEP, also called neprilysin. (EC 3.4.24.11). NEP is a membrane-bound peptidase which plays an important role in nervous and peripheral tissues as it regulates the activity of peptide agonists by catalyzing their proteolysis and removal from membrane receptors. The inhibition of NEP by sialorphin could potentially result in a longer binding time of peptide agonists to their receptors. One of the most important groups of peptide receptors are G-protein coupled receptors (GPCR), which are a superfamily of over 800 proteins estimated to be encoded by the human genome9. The GPCRs function in the regulation of transmission of signals from the cell membrane to the interior of cells. Defects in their regulation can result in uncontrolled stimulation of cellular processes, resulting in diseases such as blindness, obesity, inflammation, depression, asthma and hypertension10. Cells have evolved several mechanisms to prevent over-stimulation of GPCR signaling pathways. For example long term exposure of cells to GPCR agonists results in down-regulation of GPCR expression through altered rates of receptor degradation and synthesis11.
We hypothesized that decreased levels of Vcsa1 and consequent unregulated activity of NEP would result in shortened binding times of peptide agonists to their receptors causing compensatory changes in their expression. To test this hypothesis, we used siRNA technology to knock-down in vitro expression of Vcsa1 in corporal smooth muscle (CSM) cells and microarray and RT-PCR to determine the effect on GPCR gene expression. In addition, we confirmed these results in vivo by using a rat model. ED was induced by bilateral transection of the cavernous nerve, and the tissues were then analyzed for Vcsa1 and GPCR gene expression. Bilateral transaction of the cavernosal nerve has previously been shown to cause decreased Vcsa1 expression in corporal tissue 1, 2 in rats.
2.0 Materials and Methods
2.1 Animals
Rat CSM cells were isolated from freshly excised corporal tissues from Fischer F-344 rats as previously described 12 and grown in DMEM containing 10% FBS media. In addition, six 120-day old Fischer F-344 rats underwent surgical bilateral transection of the cavernous nerve (CN) as previously described1, 2. As controls, an additional six animals underwent sham operations without CN transection. After 9 days animals were killed by placing them in a CO2 chamber, and the corpora were harvested and flash frozen in liquid nitrogen and stored at −70°C until RNA preparation. These protocols are approved by the Animal Use Committee at the Albert Einstein College of Medicine.
2.2 Vcsa1-siRNA
siRNA that would target Vcsa1 (Vcsa1-siRNA) was constructed. We found potential Vcsa1-siRNA sequences using the Ambion on-line resource (siRNA target finder: http://www.ambion.com/techlib/misc/siRNA_finder.html). The siRNA construct, Vcsa1-siRNA, was synthesized using the Silencer siRNA Construction Kit (Ambion, Foster City, CA) following the manufacturer’s instructions. The following template oligonucleotides for the siRNAs were used: antisense, 5′-AATGGTGGACAAATAGGAGTACCTGTCTC-3′; sense, 5′-AATACTCCTATTTGTCCACCACCTGTCTC-3′.
2.3 NEP-siRNA
NEP-siRNA (smartpool) was obtained from Dharmacon Research, Inc. (Lafayette, CO). It was composed of four Smartselection designed siRNAs targeting the NEP gene with the following sequences: (1) sense, 5′-GCAGAAAUCAGAUCGUCUUUU-3′; antisense, 5′-AAGACGAUCCUGAUUUCUGCUU-3′; sense, (2) 5′-GAACAAACAUAUGGUACUUUU-3′; antisense, 5′-AAGUACCAUAUGUUUCUUCUU-3′; sense, (3) 5′-UAACCAAACUUAAGCCUAUUU-3′; antisense, 5′-AUAGGCUUAAGUUUGGUUAUU-3′; (4) sense, 5′-GUACGGACUUCUUCAAAUAUU-3′; antisense, 5′-UAUUUGAAGAAGUCCGUACUU-3′.
2.4 Transfection of corporal smooth muscle cells
The Vcsa1-siRNA or NEP-siRNA was used to knock-down expression of the Vcsa1 or the NEP gene, respectively, in rat CSM cells in vitro. siRNA transfections were carried out in 10 cm plates by using siPORT™ Lipid (Ambion, Foster City, CA) according to the manufacturer’s instruction. For controls, cells were treated under the same conditions without the inclusion of siRNA. Cells were also treated for 72 hours by addition of 1 μM phosphoramidon (Sigma-Aldrich, St. Louis, MO) to the cell culture media.
2.5 Gene Expression by Microarray analysis
RNA was isolated from rat CSM cells treated with 10 nM Vcsa1-siRNA or 25 nM NEP-siRNA and controls using the RNeasy kit (Qiagen) according to the manufacturer’s instruction. The RNA was used to perform microarray analysis of global gene expression using the RGU34A affymetrix microarray. We performed a total of 5 microarray analyses with 2 chips on control CSM cells and 3 chips on rat CSM cells treated with Vcsa1-siRNA. Gene expression comparison of the Vcsa1-siRNA treated cells with controls was performed using AffylmGUI software, available from www.bioconductor.org as previously described13. Lists of significantly changed genes were analyzed by the Expression Analysis Systematic Explorer (EASE, available from the NIH website: http://david.abcc.ncifcrf.gov/ease/ease.jsp).
2.6 Quantitative RT-PCR
RNA from cells was prepared as described above. Methods for the isolation of RNA from corporal tissue and quantitative RT-PCR have been described previously2. PCR reactions for all samples were performed in 96-well plates with 50ng of cDNA, 100 nM primers and 12.5μL of Sybr Green (PE Applied Biosystems, Warrington, United Kingdom) in a 25μL reaction volume. The cycling condition was programmed as follows: activation of Sybr Green DNA polymerase at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, annealing/extension at 60°C for 1 min. The primers used in these studies are shown in Table 1.
Table 1.
Oligonucleotide primers used in the quantitative PCR assay
| Gene | Primers |
|---|---|
| Gabra4 | Forward: 5′-ATGACAACAGACTGCGTCCTGGAT-3′ |
| Reverse: 5′-CGTTTAAACAAGTCGCCAGGCACA-3′ | |
| Ccr2 | Forward: 5′-CCAGTGTGAAGCAAATTGGAGCTTGG-3′ |
| Reverse: 5′-AGATGGCCAGGTTGAACAGGTAGA-3′ | |
| Mtnr1a | Forward: 5′-TGGTCATCCTGTCTGTGTATCGCA-3′ |
| Reverse: 5′-CAGCGCCAAGGGAAATGGGTAAAT-3′ | |
| Celsr3 | Forward: 5′-TGCTACTGGGCCAAGAGCTCAAAT-3′ |
| Reverse: 5′-TTTGCCTACCTACTCGGAGCTTCA-3′ | |
| Agtr2 | Forward: 5′-TTTGCTGCCACCAGCAGAAACATC-3′ |
| Reverse: 5′-TATCTGCCGGTTTGTGTGAGCAGT-3′ | |
| Adcyap1r1 | Forward: 5′-TGCATCTTCAAGAAGGAGCAAGCC-3′ |
| Reverse: 5′-ACCTACTTGAGCTGGCTTCCAACA-3′ | |
| Oprm1 | Forward: 5′-ACCAGTACACTGCCCTTTCAGAGT-3′ |
| Reverse: 5′-TGCAGAGGGTGAATATGCTGGTGA-3′ | |
| Vcsa1 | Forward: 5′-GAGGGTGTCAGAGGCCC-3′ |
| Reverse: 5′-GAGCAGTTAGCTGCCACTGATA-3′ | |
| NEP | Forward: 5′-TTTCTGTGCTCGTCTTGCTCCTGA-3′ |
| Reverse: 5′-TGACATTGCGTTTCAACCAGCCTC-3′ | |
| GAPDH | Forward: 5′-GCCGCCTGCTTCACCACCTTCT-3′ |
| Reverse: 5′-GCATGGCCTTCCGTGTTCCTACC-3′ |
3.0 Results
Transfection of CSM cells with 10nM Vcsa1-siRNA or with siPORT ™ transfection agent alone (mock treated controls) were carried out in 10 cm plates in DMEM media containing 10% Fetal Bovine Serum (FBS). Sixty hours post-transfection the Vcsa1 gene expression was determined using quantitative PCR. Treating CSM Vcsa1-siRNA gave >87% reduction in Vcsa1 gene expression in the cultured rat CSM cells compared to controls. The same extracted RNA was used to perform microarray analysis of global gene expression using the RGU34A affymetrix microarray. A list of significantly changed genes (determined by the B-statistic) was generated using the AffylmGUI software, available from www.bioconductor.org 13. We then analyzed the biological function of the significantly changed genes using expression analysis systematic explorer (EASE14). In Table 2 we show identified groups of genes where the EASE score is less than 0.05. Our analysis showed highly significant overrepresentation of genes involved in G-protein coupled receptor (GPCR) activity.
Table 2.
EASE analysis of gene functional characteristics over-represented in the changed gene lists after knockdown of expression of Vcsa1.
| System | Category | LH | LT | PH | PT | EASE |
|---|---|---|---|---|---|---|
| cellular component | integral to membrane | 23 | 37 | 783 | 1932 | 0.0095 |
| molecular function | receptor activity | 14 | 43 | 375 | 2367 | 0.0264 |
| molecular function | G-protein coupled receptor activity | 8 | 43 | 159 | 2367 | 0.0348 |
| molecular function | signal transducer activity | 18 | 43 | 577 | 2367 | 0.0458 |
EASE analysis (http://david.niaid.nih.gov/david/ease.htm, (22)) of genes with changed expression following knock-down of Vcsa1 using siRNA at 25 nM. LH=List Hits (number of genes in the gene list that belong to the gene category. LT= List Total (number of genes in the gene list) PH=Population Hits (number of genes in the total group of genes assayed that belong to the specific gene category). PT= Population Total number of genes in the total group of genes assayed that belong to any category within the system).
In Table 3 we show the 43 assayed genes on the RGU34A microarray that fall into the group defined as having GPCR activity. The fold change in gene expression indicates up-regulation in the Vcsa1-siRNA treated cells compared to the controls. The B-statistic indicates the significance of the change in gene expression; a score >1 indicates a statistically significant change in gene expression compared to untreated controls. Not all the GPCR changes in expression have peptide agonists, which is surprising considering that the protein product of Vcsa1, sialorphin, is an endogenous peptidase inhibitor.
Table 3.
G-coupled protein receptors present in corporal smooth muscle cells (detected by microarray analysis) and change in expression when treated with Vcsa1-NEP.
| Symbol | Gene | B-statistic | Fold Change |
|---|---|---|---|
| Gabra4 | gamma-aminobutyric acid (GABA-A) receptor, subunit alpha 4 | 12.41037488 | 20.68691607 |
| Ccr2 | chemokine (C-C motif) receptor 2 | 14.20093557 | 17.19107659 |
| Hcrtr2 | hypocretin (orexin) receptor 2 | 12.51012791 | 15.86347807 |
| Mtnr1a | melatonin receptor 1A | 11.43199341 | 11.21879566 |
| Celsr3 | cadherin EGF LAG seven-pass G-type receptor 3 | 9.0069665 | 10.29157574 |
| Ccr5 | chemokine (C-C) receptor 5 | 10.67853803 | 9.966672355 |
| Agtr2 | angiotensin II receptor, type 2 | 7.787873351 | 9.154239457 |
| Adcyap1r1 | adenylate cyclase activating polypeptide 1 receptor 1 | 11.69137911 | 8.768922888 |
| Oprm1 | opioid receptor, mu 1 | 9.947061929 | 7.321373484 |
| Grm2 | glutamate receptor, metabotropic 2 | 9.432481922 | 6.876953784 |
| Calcrl | calcitonin receptor-like | 10.07497323 | 6.42708125 |
| Fshr | follicle stimulating hormone receptor | 8.764853382 | 6.34367168 |
| Nxph1 | neurexophilin 1 | 9.195471437 | 5.415030946 |
| Arl3 | ADP-ribosylation factor-like 3 | 8.571346447 | 4.960725826 |
| Gpr24 | G protein-coupled receptor 24 | 5.88419255 | 4.737365123 |
| Ecel1 | endothelin converting enzyme-like 1 | 6.804210975 | 4.468540643 |
| C3ar1 | complement component 3a receptor 1 | 8.560846324 | 4.408652995 |
| Gabrr2 | gamma-aminobutyric acid A receptor, rho 2 | 8.652629815 | 4.22493427 |
| Chrm5 | cholinergic receptor, muscarinic 5 | 3.675843876 | 3.866131237 |
| Gabrg2 | gamma-aminobutyric acid A receptor, gamma 2 | 7.981849786 | 3.491850763 |
| Mas1 | MAS1 oncogene | 6.703196086 | 2.915382323 |
| Grb14 | growth factor receptor bound protein 14 | 0.279054051 | 2.846778548 |
| Sstr1 | somatostatin receptor 1 | 5.813340243 | 2.811720785 |
| Vipr1 | vasoactive intestinal peptide receptor 1 | 5.854036807 | 2.573352521 |
| Ghrh | growth hormone releasing hormone | 3.464521913 | 2.516117866 |
| Grik2 | glutamate receptor, ionotropic, kainate 2 | 4.558269023 | 2.430048197 |
| Drd2 | dopamine receptor 2 | 6.328035066 | 2.415420604 |
| Adrb2 | adrenergic receptor, beta 2 | 4.83859972 | 2.291419935 |
| F2r | coagulation factor II (thrombin) receptor | 4.981961453 | 1.888623241 |
| Htr5a | 5-hydroxytryptamine (serotonin) receptor 5A | 5.022532023 | 1.801031313 |
| Chrm2 | cholinergic receptor, muscarinic 2 | 1.017576296 | 1.446614704 |
| Gria3 | glutamate receptor, ionotropic, AMPA3 (alpha 3) | 3.123862632 | 1.44637896 |
| Gpr20 | G protein-coupled receptor 20 | 1.776954221 | 1.410922562 |
| Chrm3 | cholinergic receptor, muscarinic 3 | 3.348453846 | 1.339036933 |
| Htr7 | 5-hydroxytryptamine (serotonin) receptor 7 | 0.47710741 | 1.18283939 |
| Ptgdr | prostaglandin D receptor | 0.239210351 | 0.998857706 |
| Adrb3 | adrenergic receptor, beta 3 | 1.805930858 | 0.984280774 |
| Drd4 | dopamine receptor 4 | 2.043147208 | 0.949623286 |
| Tacr2 | tachykinin receptor 2 | 0.981167624 | 0.931291478 |
| P2ry2 | purinergic receptor P2Y, G-protein coupled 2 | 0.604120257 | 0.882479154 |
| Gng8 | guanine nucleotide binding protein (G protein), gamma 8 subunit | 1.150735789 | 0.813719316 |
| Nos2 | nitric oxide synthase 2, inducible | 0.134838156 | 0.704032284 |
| Gnas | GNAS complex locus | 0.461991262 | 0.632688694 |
A complete list of the G-coupled protein receptors with changed expression in following knock-down of expression of Vcsa1 using siRNA. RNA was prepared from treated and untreated cells, reverse transcribed to cDNA, labeled with biotin and fragmented according to protocols developed by Affymetrix (www.affymetrix.com). Subsequent hybridization to microarray chips was performed at AECOM microarray core facilities using an Affymetrix Fluidics Station. Analysis of the data was performed AffylmGUI, available from www.bioconductor.org. Briefly the steps were to visually check for artifacts on the Chip (and exclude outlying chips from further analysis), normalize raw data using robust multi-array averaging (RMA) and calculate the fold change in level of gene expression by comparing experiment and control groups. Significant change was determined by the B statistic (which is the log odds of differential expression, which takes into account the variability between replicate arrays).
Primers were designed to determine the expression for seven of the ten most up-regulated GPCR genes identified by microarray, as shown in Table 1. Rat CSM cells growing in vitro were treated with 10 nM Vcsa1-siRNA, RNA was isolated and quantitative-RT-PCR performed to determine the level of expression of specific GPCR genes relative to untreated controls. The results (see Figure 1) confirm that the GPCR transcripts identified as up-regulated in the microarray analysis are also up-regulated when expression is determined by quantitative-PCR. The fold-change in up-regulation of expression of GPCR in treated compared to controls is somewhat less (approximately 4-fold) than that seen with microarray analysis. We surmised that the up-regulation of GPCR was a direct response to the down-regulation of sialorphin expression after the treatment of cells with Vcsa1-siRNA. With less sialorphin produced by the cells, there would be less inhibition of NEP. Therefore, agonists present in the serum would be more rapidly removed from their receptors, with a consequent decrease in the activity of the down-stream signaling pathways. In order to compensate for decreased activation of signaling pathways, we hypothesize that cells up-regulate expression of GPCR.
Fig. 1.
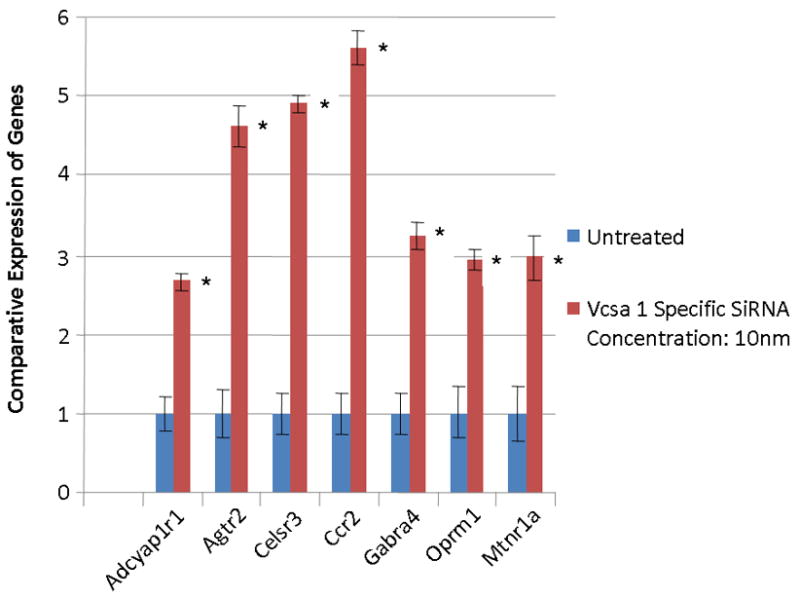
The expression of GPCR in corporal cells in vitro following transfection of 10 nM Vcsa1-siRNA for 60 hours in DMEM containing 10% FBS. Expression of the various genes were determined by quantitative PCR, normalized to GAPDH, and the untreated cells used as the calibrator, essentially as described in 2. The experiment was repeated three times, and the expression of the GPCR determined in triplicate. In all GPCR genes investigated the gene is significantly up-regulated compared to control (significance is determined by the students t-test, * = P <0.05). Adcyap1r1= adenylate cyclase activating polypeptidereceptor subtype 1r1, Agtr2 angiotensin II receptor, type 2, Celsr3=cadherin EGF LAG seven-pass G-type receptor 3, Ccr2=chemokine (C-C motif) receptor 2, gamma-aminobutyric acid (GABA-A) receptor, subunit alpha 4, Oprm1=opioid receptor, mu 1, Mtnr1a=melatonin receptor 1A.
We hypothesized that inhibiting NEP using phosphoramidon 15 would result in the down-regulation of GPCR expression as opposed to their up-regulation following treatment of cells with Vcsa1-siRNA. This is because NEP inhibition by phosphoramidon would result in longer binding times of agonist to GPCR, and would result in compensatory changes in cells that would down-regulate GPCR gene expression to prevent over activity of the G-protein downstream signaling pathways. As seen in Figure 2, the majority of GPCR are down-regulated by the presence of 1μM phosphoramidon in the culture media. The exception was Celsr3, a cadherin EGF LAG seven-pass G-type receptor, which was up-regulated. This may be a result of over-lapping, but not identical, specificities in inhibiting cell surface expressed peptidases by sialorphin compared to phosphoramidon.
Fig. 2.
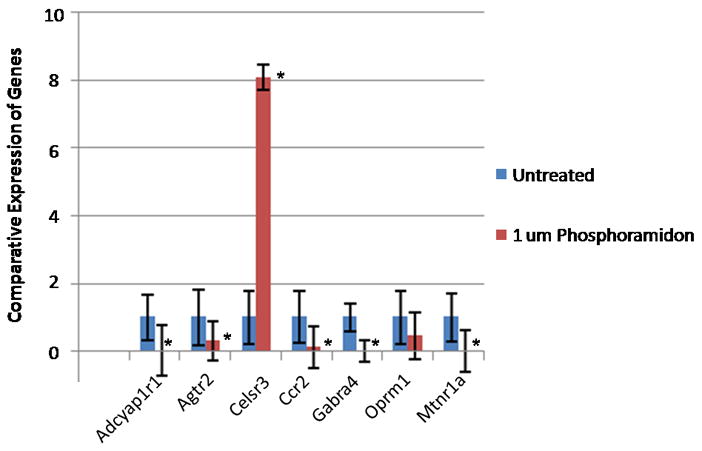
The expression of GPCR in corporal cells in vitro following treatment with 1 μM phosphoramidon. The experiment was repeated three times, and the expression of the GPCR determined in triplicate (significance is determined by the students t-test, * = P <0.05).. In all GPCR genes investigated the gene is significantly down-regulated compared to control with the exception of Celsr3 where the gene is significantly up-regulated. (Gene abbreviations are the same as in Fig 1).
We hypothesize that the effect on expression of GPCR expression by sialorphin and phosphoramidon is mediated through their affect on the NEP. In order to directly demonstrate that NEP directly influences GPCR expression, we treated CSM cells with 25 nM NEP-siRNA and compared expression of the GPCR with untreated controls. The down-regulation of NEP, which would lead to extended binding times of agonists on GPCR and activation of signaling pathways, should result in a compensatory down regulation of GPCR. Initial experiments demonstrated that treatment of CSM cells in vitro with 25 nM NEP-siRNA resulted in >80% reduction of NEP expression. As seen in Figure 3, most of the GPCR genes investigated were significantly reduced in expression with the exception of Ccr2, which was not significantly changed in expression, and Celsr3, which was significantly up-regulated with the knockdown in expression of NEP.
Fig. 3.
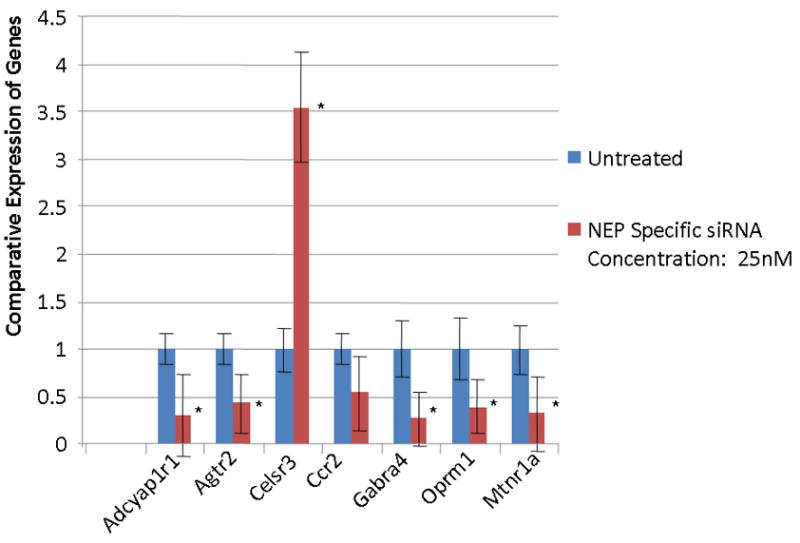
The expression of GPCR in corporal cells in vitro following transfection of 25 nM NEP-siRNA for 60 hours in DMEM. Expression of the various genes were determined by quantitative PCR, normalized to GAPDH, and the untreated cells used as the calibrator, essentially as described in 2. The experiment was repeated three times, and the expression of the GPCR determined in triplicate (significance is determined by the students t-test, * = P <0.05). In all GPCR genes investigated the gene is significantly down-regulated compared to control. An exception is Celsr3 where the gene is significantly up-regulated and Ccr2 where there was no significant change in expression compared to controls. (Gene abbreviations are the same as in Fig 1).
It has previously been demonstrated that animal models for ED have decreased Vcsa1 expression in corporal tissue 1, 2. The decrease in Vcsa1 expression following CN transection was previously reported as maximal at one week, after which Vcsa1 expression slowly increased 1. Therefore, we evaluated if changes in the expression of Vcsa1 one week after bilateral transection of CN were accompanied with changes in GPCR expression. As previously reported, there was >90% decrease in Vcsa1 expression in animals that underwent bilateral CN transection. Quantitative RT-PCR demonstrated that the majority of GPCR genes that were up-regulated in in vitro knockdown expression of Vcsa1 (Figure 1) were also up-regulated in animals with ED caused by bilateral CN transection (Figure 4).
Fig. 4.
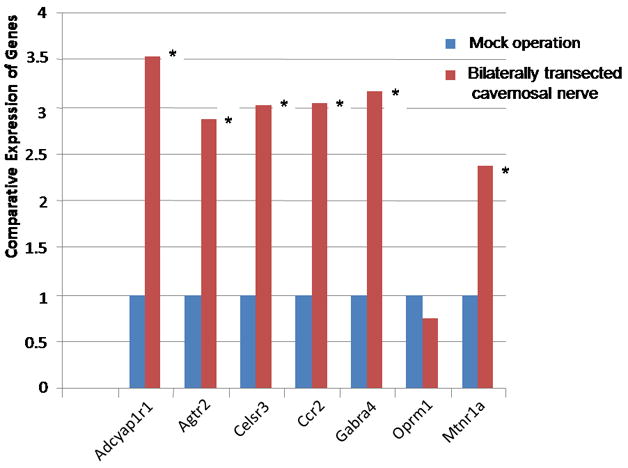
The expression of GPCR in corporal tissue following bilaterally ligation of the cavernous nerve for 9 days compared to mock operated animals. Expression of the various genes were determined by quantitative PCR, normalized to GAPDH, and the untreated cells used as the calibrator, essentially as described in 2. The results are an average of six animals, each reading performed in duplicate. In all GPCR genes investigated (except Oprm1) the gene is significantly up-regulated compared to mock treated animals (significance is determined by the students t-test, * =P <0.05). An exception is Celsr3 where the gene is significantly up-regulated (P <0.05). (Gene abbreviations are the same as in Fig 1).
4.0 Conclusion and Discussion
Our results demonstrate that the knock-down of Vcsa1 expression in vitro causes significant up-regulation of GPCR gene expression as an ontological group. The effect is opposite to the effect of treating cells with a synthetic NEP inhibitor, phosphoramidon, or treating cells with NEP-siRNA. The gene product of Vcsa1, sialorphin, very efficiently protects peptide agonists from breakdown by cell surface endopeptidase in vitro 5. Therefore, we believe that in Vcsa1-siRNA-treated cells which have increased NEP activity (as a result of the down-regulation of sialorphin) agonists have shorter binding times to GPCR. This leads to a decrease in the down-stream signaling pathways. In order to compensate for the reduced activity of these signaling pathways, cells increase the expression of GPCR genes. Conversely, treatments which reduce NEP activity (eg. adding phosphoramidon to culture media or treating cells with NEP-siRNA) would prolong the activity of agonists at their receptors, activating downstream signaling pathways, and resulting in a compensatory down-regulation of GPCR. In vivo we have shown that the previously reported down-regulation of Vcsa1, which occurs in several rat models of ED, is accompanied by the up-regulation of GPCR gene expression, suggesting that similar mechanisms of GPCR regulation may operate in rat corporal tissue. Although the changes in Vcsa1 and GPCR expression correlate with the in vitro Vcsa1-siRNA knock-down experiments, CN transection has severe effects on the anatomy of the corpora such as penile shrinkage and apoptosis of corporal smooth muscle cells 16. The definitive role of Vcsa1 in modulating GPCR expression in vivo may only be determined with the development of viral vectors expressing Vcsa1-inhibitory RNA or Vcsa1−/− knock-out mice. The interactive effects of GPCR, NEP and sialorphin are summarized in Fig 5. Since the effect on expression is not restricted to GPCR that have peptide agonists, the effect is probably mediated through downstream G-protein activated pathways.
Fig. 5.
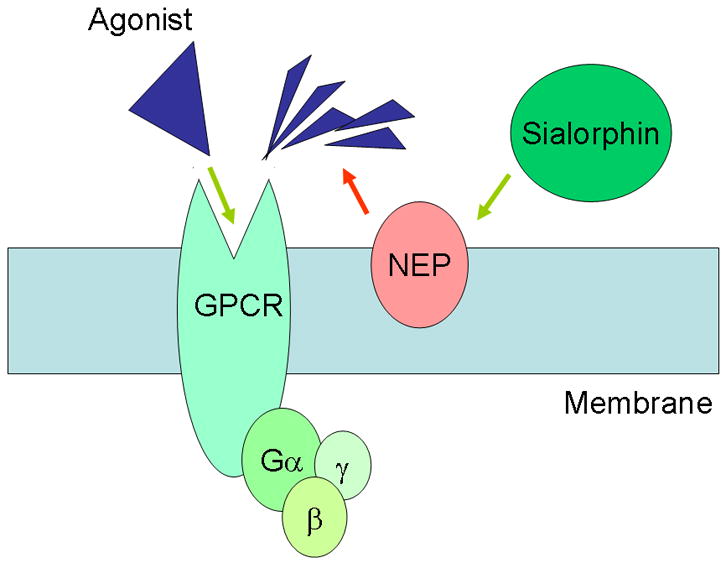
An agonist binds to G-protein coupled receptors to activate Gμ subunit. Neutral endopeptidase acts on the agonist to remove it. The action of sialorphin is to prolong agonist stimulation by preventing the action of NEP.
We have demonstrated that sialorphin (and expression of the human homologous gene, hSMR3A) can modulate erectile physiology 2, 3. The work presented here suggests that one possible mechanism by which the protein products of the Vcsa1 family of genes are implicated in erectile physiology involves the regulation of GPCR gene expression in corporal tissue. One member of the GPCR gene family, melanocortin receptor 4 or MC4R, has been shown to modulate erectile function in mice17, and several agonists of MC4R are under investigation as potential erectogenic drugs18. So far, our microarray studies have shown no significant difference in MCR4 expression in Vcsa1-siRNA treated corporal cells in vitro. A number of GPCR are changed in expression with the onset of ED, indicating that they are potential targets for the development of clinically active erectogenic agents. Vcsa1 and hSMR3A have been suggested as markers for ED2, 3. Therefore, measuring changes in GPCR expression may also be useful in determining the degree of ED and the predicting the efficacy of treatment.
It is possible that ED, which results in the down-regulation of Vcsa12 (or hSMR3A in human patients3), and therefore potentially circulating levels of their peptide products, can result in changes in GPCR expression in tissues distal to corporal tissue, such as other vascular tissues with consequent pathology. Synthetic NEP inhibitors, such as phosphoramidon and thiorphan have been shown to have physiological effects on the vascular, cardiovascular and renal function in rats 15. It is therefore feasible that several of the vascular problems associated with diabetic patients, such as hypertension and cardiovascular disease, are related to down-regulation of opiorphin and the consequent up-regulation GPCR expression. Recently a close correlation between erectile dysfunction (ED) and cardiovascular disease has been reported, and patients reporting ED are referred for screening for underlying cardiovascular disease 19. It is assumed that this correlation is due to common mechanisms for the development of disease in the endothelial cells of blood vessels in the penis and heart20. However, our work raises the possibility that other mechanisms may also be important and that the down-regulation of expression of human sialorphin homologues in the penis, and thereby circulating levels of sialorphin, may potentially modulate vascular tissue physiology through alterations in GPCR activity.
Acknowledgments
This work was supported by grants awarded by the NIH/NIDDK to Kelvin P. Davies (R21DK079594 and R01DK077665) and Arnold Melman (P01DK060037). We thank Alison Duncan and Dr. Sarah Collins for proof reading the manuscript.
Abbreviations
- CN
Cavernous nerve
- GPCR
G-protein coupled receptor
- ED
erectile dysfunction
- NEP
neutral endopeptidase
- Vcsa1
variable coding sequence protein A1
- SMR3A
sub-mandibular protein
- RT-PCR
reverse transcription-polymerase chain reaction
- Adcyap1r1
adenylate cyclase activating polypeptidereceptor subtype 1r1 Agtr2 angiotensin II receptor, type 2
- Celsr3
cadherin EGF LAG seven-pass G-type receptor 3
- Ccr2
chemokine (C-C motif) receptor 2, gamma-aminobutyric acid (GABA-A) receptor, subunit alpha 4
- Oprm1
opioid receptor, mu 1
- Mtnr1a
melatonin receptor 1A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.User HM, Zelner DJ, McKenna KE, McVary KT. Microarray analysis and description of SMR1 gene in rat penis in a post-radical prostatectomy model of erectile dysfunction. J Urol. 2003;170:298. doi: 10.1097/01.ju.0000060882.75475.5a. [DOI] [PubMed] [Google Scholar]
- 2.Tong Y, Tar M, Davelman F, Christ G, Melman A, Davies KP. Variable coding sequence protein A1 as a marker for erectile dysfunction. BJU Int. 2006;98:396. doi: 10.1111/j.1464-410X.2006.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong Y, Tar M, Monrose V, DiSanto M, Melman A, Davies KP. hSMR3A as a marker for patients with erectile dysfunction. J Urol. 2007;178:338. doi: 10.1016/j.juro.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wisner A, Dufour E, Messaoudi M, Nejdi A, Marcel A, Ungeheuer MN, et al. Human Opiorphin, a natural antinociceptive modulator of opioid-dependent pathways. Proc Natl Acad Sci U S A. 2006;103:17979. doi: 10.1073/pnas.0605865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rougeot C, Messaoudi M, Hermitte V, Rigault AG, Blisnick T, Dugave C, et al. Sialorphin, a natural inhibitor of rat membrane-bound neutral endopeptidase that displays analgesic activity. Proc Natl Acad Sci U S A. 2003;100:8549. doi: 10.1073/pnas.1431850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rougeot C, Rosinski-Chupin I, Njamkepo E, Rougeon F. Selective processing of submandibular rat 1 protein at dibasic cleavage sites. Salivary and bloodstream secretion products. Eur J Biochem. 1994;219:765. doi: 10.1111/j.1432-1033.1994.tb18556.x. [DOI] [PubMed] [Google Scholar]
- 7.Davies KP, Tar M, Rougeot C, Melman A. Sialorphin (the mature peptide product of Vcsa1) relaxes corporal smooth muscle tissue and increases erectile function in the ageing rat. BJU Int. 2007;99:431. doi: 10.1111/j.1464-410X.2006.06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messaoudi M, Desor D, Nejdi A, Rougeot C. The endogenous androgen-regulated sialorphin modulates male rat sexual behavior. Horm Behav. 2004;46:684. doi: 10.1016/j.yhbeh.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002;520:97. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 10.Smit MJ, Vischer HF, Bakker RA, Jongejan A, Timmerman H, Pardo L, et al. Pharmacogenomic and Structural Analysis of Constitutive G Protein-Coupled Receptor Activity. Annu Rev Pharmacol Toxicol. 2007;47:53. doi: 10.1146/annurev.pharmtox.47.120505.105126. [DOI] [PubMed] [Google Scholar]
- 11.Bohm SK, Grady EF, Bunnett NW. Regulatory mechanisms that modulate signalling by G-protein-coupled receptors. Biochem J. 1997;322 (Pt 1):1. doi: 10.1042/bj3220001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang HZ, Day N, Valcic M, Hsieh K, Serels S, Brink PR, et al. Intercellular communication in cultured human vascular smooth muscle cells. Am J Physiol Cell Physiol. 2001;281:C75. doi: 10.1152/ajpcell.2001.281.1.C75. [DOI] [PubMed] [Google Scholar]
- 13.Hipp JD, Davies KP, Tar M, Valcic M, Knoll A, Melman A, et al. Using gene chips to identify organ-specific, smooth muscle responses to experimental diabetes: potential applications to urological diseases. BJU Int. 2007;99:418. doi: 10.1111/j.1464-410X.2007.06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marton Z, Pataricza J, Krassoi I, Varro A, Papp JG. NEP inhibitors enhance C-type natriuretic peptide-induced relaxation in porcine isolated coronary artery. Vascul Pharmacol. 2005;43:207. doi: 10.1016/j.vph.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 16.User HM, Hairston JH, Zelner DJ, McKenna KE, McVary KT. Penile weight and cell subtype specific changes in a post-radical prostatectomy model of erectile dysfunction. J Urol. 2003;169:1175. doi: 10.1097/01.ju.0000048974.47461.50. [DOI] [PubMed] [Google Scholar]
- 17.Gantz I, Fong TM. The melanocortin system. Am J Physiol Endocrinol Metab. 2003;284:E468. doi: 10.1152/ajpendo.00434.2002. [DOI] [PubMed] [Google Scholar]
- 18.Martin WJ, MacIntyre DE. Melanocortin receptors and erectile function. Eur Urol. 2004;45:706. doi: 10.1016/j.eururo.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Thompson IM, Tangen CM, Goodman PJ, Probstfield JL, Moinpour CM, Coltman CA. Erectile dysfunction and subsequent cardiovascular disease. Jama. 2005;294:2996. doi: 10.1001/jama.294.23.2996. [DOI] [PubMed] [Google Scholar]
- 20.Kirby M, Jackson G, Simonsen U. Endothelial dysfunction links erectile dysfunction to heart disease. Int J Clin Pract. 2005;59:225. doi: 10.1111/j.1742-1241.2005.00453.x. [DOI] [PubMed] [Google Scholar]


