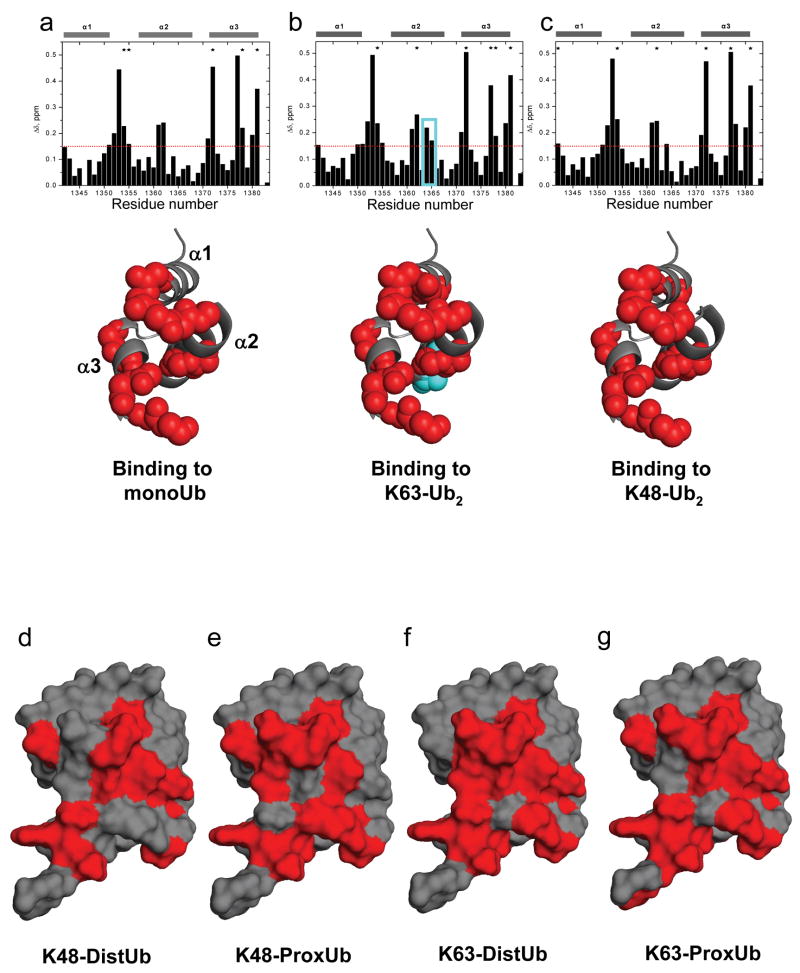
Figure 2.
CSP mapping of the interactions of Ede1 UBA with monoUb, K63-polyUb and K48-polyUb reveals no linkage-specific mode of interaction. Amide CSPs at titration endpoints are shown as a function of residue number for the titrations of 15N-Ede1 UBA with (a) monoUb, (b) K63-Ub2, and (c) K48-Ub2 (upper panels). Residues that were significantly perturbed upon binding (Δδ>0.15 ppm, or signal attenuation >60%) are mapped to the Ede1 UBA structure12 (2g3q.pdb) in red spheres below each plot. E1364 and K1365 are shown in cyan in (b). Likewise, Ub2 molecules were segmentally labeled with 15N and titrated with unlabeled Ede1 UBA. Ub residues that were significantly perturbed (Δδ>0.1 ppm, or signal attenuation >60%) are mapped to the surface of Ub12 (2g3q.pdb) in red for the (d) distal and (e) proximal K48-linked Ubs, and for the (f) distal and (g) proximal K63-linked Ubs. The CSP values are shown in Supplemental Figure 3, along with 15N monoUb CSPs, which are highly similar to both polyUb measurements.