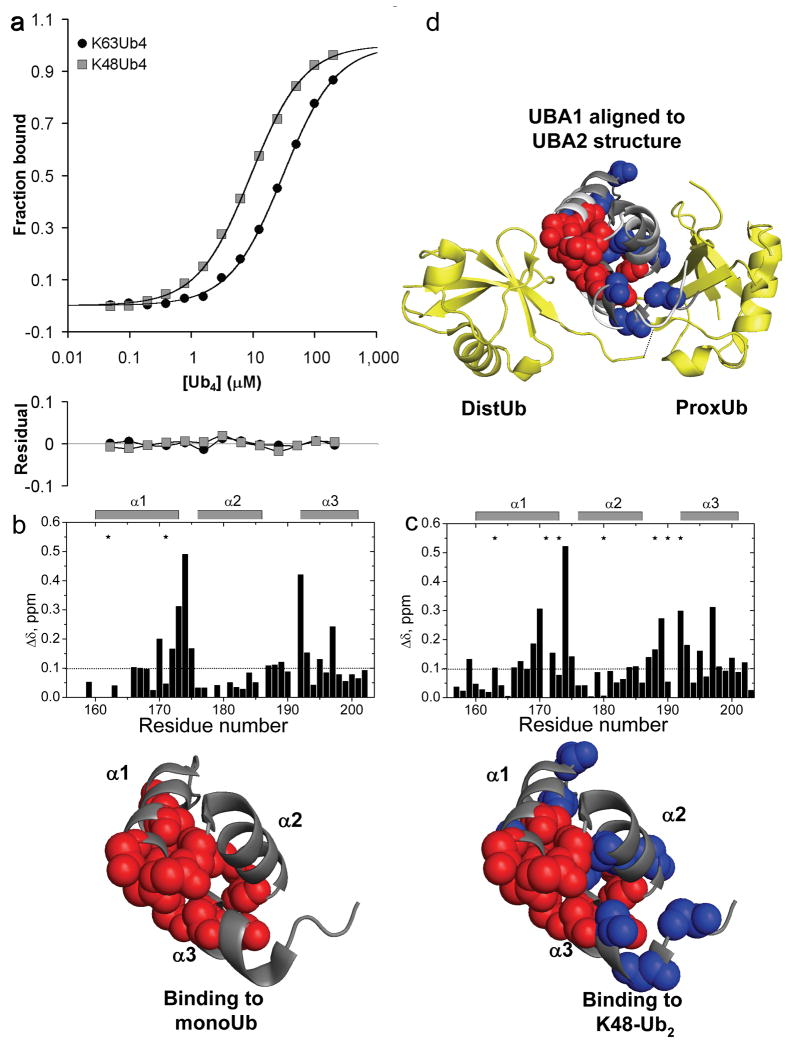
Figure 5.
hHR23A-UBA1 is a K48-selective UBA domain. (a) Fluorescence anisotropy binding data for hHR23A-UBA1 interacting with K63-Ub4 or K48-Ub4 indicate a preference for K48-polyUb. CSP mapping was used to identify the UBA1 surface responsible for binding to (b) monoUb and (c) K48-Ub2. Upper panels show the amide CSPs as a function of 15N-UBA1 residue number. Residues that were significantly affected by binding (CSP Δδ>0.10 ppm, or signal attenuation >60%) are mapped to the UBA1 structure20 (1IFY.pdb) below (spheres). Blue spheres indicate the residues that were only perturbed upon K48-Ub2 binding. UBA1 interacts with K48-Ub2 with an expanded set of residues in a configuration that is similar to the linkage-specific binding of UBA2 from the same protein. (d) To show this similarity, we aligned UBA1 (contacts indicated as before by spheres) to the UBA2 coordinates from its bound complex with K48-Ub2 (1ZO6.pdb)13. Ub2 is in yellow ribbons; UBA2 is in white ribbons; the aligned UBA1 is in dark gray ribbons. The putative interface of UBA1 with the distal Ub is similar to the monoUb interface (red spheres), whereas additional residues on the other side of UBA1 (blue spheres) that are specifically perturbed in the K48 interaction could make linkage-specific interactions with the isopeptide region and proximal Ub.