Fig. 1.
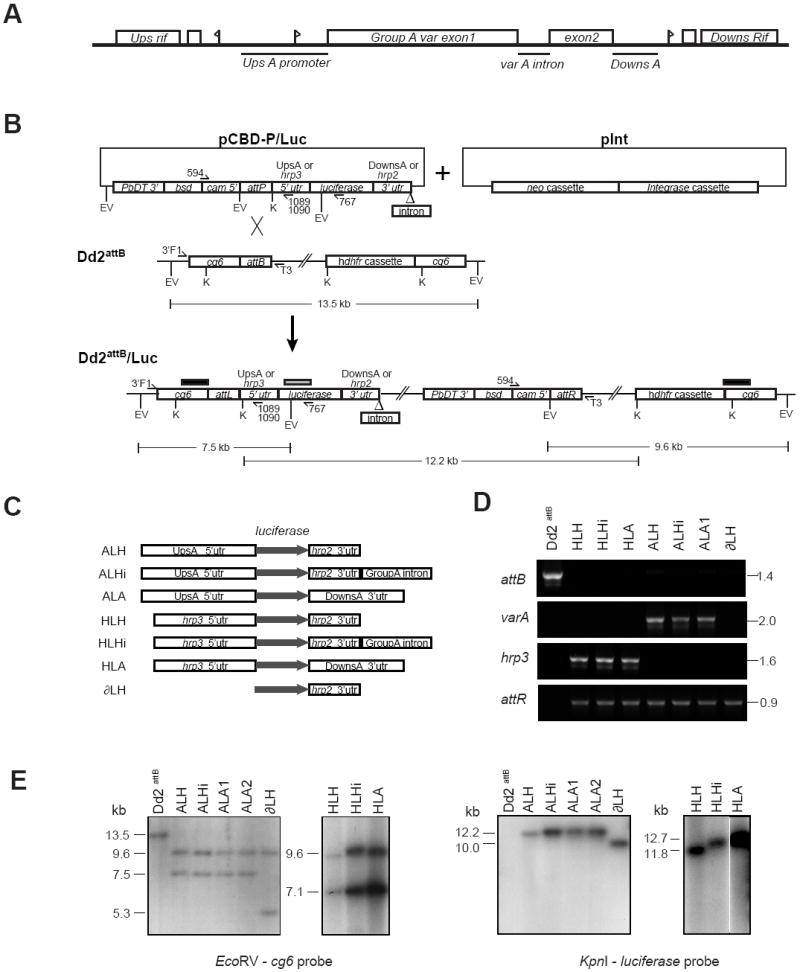
Generation and molecular characterization of luciferase reporter cassettes integrated into the Plasmodium falciparum Dd2attB parasite line. (A) Schematic of the genomic layout of the Group A var gene PFD1235w (previously named AL010226) in the P. falciparum 3D7 strain. Flags represent putative transcription start sites. Bars represent DNA regions chosen for analysis. The diagram is not drawn to scale. (B) Schematic of the integration of a generic luciferase reporter cassette-bearing pCBD-P plasmid (containing the attP element) into the attB locus of the Dd2attB parasite line. The attB docking site was engineered into the cg6 gene by homologous recombination (Nkrumah et al., 2006). Co-transfection of pCBD-P plasmids with the integrase-encoding plasmid pInt resulted in site-specific integration of the pCBD-P plasmid into the genome, flanked by attL and attR sequences. Upon recombination, the luciferase cassette resided downstream of the cg6 amino-terminal region, followed by the pBluescript plasmid backbone and the bsd selectable marker cassette placed in the opposite orientation to the reporter. Restriction digest fragment sizes refer to the integration of the pCBD-P-ALH plasmid. The positions of the probes used for Southern blot hybridization are indicated as bars above the Dd2attB/Luc locus (black bar: cg6 probe; gray bar: luciferase probe). EV, EcoRV; K, KpnI. (C) Experimental design of luciferase reporter assays, depicting the cassettes subcloned into pCBD-P and integrated into Dd2attB. The grey arrow indicates the luciferase open reading frame. UpsA, the 2.15 kb 5’ untranslated region (UTR) of PFD1235w; DownsA, the 1.4 kb 3’ UTR of PFD1235w; Group A intron, the 0.74 kb PFD1235w intron. (D) PCR reactions documenting the integration of luciferase reporters into Dd2attB. attB: Amplification of the Dd2attB parent strain and designated luciferase reporter lines with primers 3’F1 (cg6-specific) and T3 (pBS plasmid backbone). varA and hrp3: Results of PCR over the attL recombination site using the primer 3’F1 and either the UpsA-specific primer 1089 (varA) or the hrp3-specific primer 1090 (hrp3). attR: PCR amplification over the attR junction using primers 594 (specific to the cam promoter driving the bsd selectable marker in all pCBD-P plasmids) and T3. The parasite line ALA2 yielded similar results (data not shown). (E) Southern blot hybridization documented the insertion of single-copy luciferase cassettes into the attB site of the Dd2attB parasite. Genomic DNA was isolated from parasite lines and digested with EcoRV or KpnI. Hybridization with a cg6 probe revealed a 13.5 kb fragment in the EcoRV-digested Dd2attB genomic DNA that was absent in all second generation attP recombinants. This band was replaced in all lines by a 9.6 kb band corresponding to the attR junction and the human dhfr cassette, and a band of variable size (due to the different promoters tested) that corresponded to the attL junction and the 5’ portion of the luciferase cassette. The smaller band sizes are as follows: ALH, ALHi and ALA cassettes: 7.5 kb; HLH, HLHi and HLA cassettes: 7.1 kb; ∂LH cassette: 5.3 kb. Hybridization with a luciferase probe showed bands consisting of the entire inserted pCBD-P plasmid, the attR junction, and the plasmid backbone of the integrated attB locus. There was no evidence of plasmid retention or multi-copy insertion, which would have been apparent as bands ~3 kb smaller in size. Sizes were as follows: ALH: 12.2 kb; ALHi: 12.9 kb; ALA: 13.1 kb; ∂LH: 10.0 kb; HLH: 11.8 kb; HLHi: 12.6 kb; and HLA: 12.7 kb. The autoradiograph of the KpnI digested genomic DNA from the hrp3 containing lines HLH, HLHi and HLA was cropped to remove intervening lanes.
