Abstract
Background
Pectobacterium carotovorum subsp. carotovorum is a phytopathogenic enterobacterium responsible for soft rot, a disease characterized by extensive maceration of the affected plant tissue. This species also produces two or more antibacterial substances called bacteriocins, which enhance its competitiveness against related rival species. However, the secretion mechanism for low-molecular-weight bacteriocin is still unknown.
Results
A mutant (flhC::Tn5) that did not secrete the low-molecular-weight bacteriocin (LMWB), Carocin S1, was generated by Tn5 insertional mutagenesis. Sequence analysis indicated that this insertion disrupted open reading frame 2 (ORF2) and ORF3 of this strain. Deletion and rescue experiments indicated that ORF2 and ORF3 were both required for extracellular LMWB secretion. The ORF2 and ORF3 sequences showed high homology with the flhD and flhC gene sequences of Pectobacterium carotovorum subsp. atroseptica, Serratia marcescens, Yersinia enterocolitica, and Escherichia coli, indicating that they likely encoded key regulatory components of the type III flagella secretion system.
Conclusion
Thus, the extracellular export of Carocin S1 by Pectobacterium carotovorum subsp. carotovorum appears to utilize the type III secretion system integral to bacterial flagella.
Background
Pectobacterium carotovorum subsp. carotovorum is a phytopathogenic enterobacterium responsible for soft rot, a disease characterized by extensive plant tissue maceration caused by a variety of secreted enzymes. The major pathogenicity determinants are an arsenal of extracellular pectinases, including several pectate lyase isozymes: pectin lyase, pectin methylesterase, and pectin polygalacturonase. In addition, a range of other degradative enzymes, such as cellulase and proteases, play equivocal roles in virulence [1]. Pectobacterium carotovorum subsp. carotovorum also produces one or more antibacterial substances called bacteriocins, which enhance their competitiveness with other related rival species [2]. The ability of this bacterial species to produce bacteriocin has been exploited in many biological control programs for the soft-rot disease of Chinese cabbage [3-5]. In view of this, identification and cloning of the gene(s) controlling bacteriocin production may facilitate the development of wider and more innovative control methods, such as the cloning of these gene(s) into Chinese cabbage, tobacco, and other susceptible plants to produce resistant cultivars.
In our previous paper, the brg gene was found to encode a regulator required for the expression of the low-molecular-weight bacteriocin (LMWB) in a strain of Pectobacterium carotovorum subsp. carotovorum [1]. The gene is homologous to hfq and encodes a protein with similar functions [1,6]. The genetic determinant encoding LMWB synthesis was designated the Carocin S1 genetic determinant, which consists of two structural genes, caroS1K (encoding killer protein) and caroS1I (immunity protein). Clear zones of inhibition around CaroS1K producer colonies are due to CaroS1K antibiotic activity. Carocin S1-associated nuclease activity has also been demonstrated [7]. The carocin S1 gene has been isolated from Pectobacterium carotovorum subsp. carotovorum 89-H-4 and functionally expressed after introduction into Pectobacterium carotovorum subsp. carotovorum Ea1068a (a non-bacteriocin-producing strain). From our previous studies, glucose, as well as SOS agents, can also induce the carocin S1 gene.
Using the same Carocin S1-producing strain of Pectobacterium carotovorum subsp. carotovorum, genes controlling the LMWB have been cloned and sequenced, and homology to the flhD/C operon demonstrated. The flhD/C operon is a regulator that activates expression of genes required for flagella assembly. However, its activity depends on environmental stimuli (e.g., cyclic AMP levels, temperature, heat shock, osmolarity, membrane biosynthesis, and H-NS protein [8]), cell division, flagella formation, and motility [9-11].
A number of Gram-negative pathogenic bacteria have evolved a specialized type III protein secretion system to deliver effector virulence proteins into host cells [12,13]. There are two types of type III secretion systems: the translocation-associated type III secretion system (T3aSS) and the bacterial flagellum type III secretion system (T3bSS). The various bacterial type III secretion systems characterized thus far all have Sec independence, ATPase dependence, presence of a hollow filamentous organelle that extends from the outer membrane, a cell-envelope-spanning secretion channel, and nine conserved proteins [14].
The bacterial flagellum type III secretion system also serves as the bacterial flagellum (a biological nanomachine with an ion-powered rotary motor). For the flagellum, the T3bSS apparatus functions to secrete components including the rod, hook, and filament subunits for extracellular assembly. The core of the flagellum is hollow, and secreted subunits polymerize at the growing end of the flagellum. A cap at the tip of the flagellum ensures efficient polymerization of secreted subunit proteins [15,16]. This secretion apparatus is just one mechanism utilized by Gram-negative plant and animal pathogens for the secretion and translocation of virulence determinants into susceptible eukaryotic cells [17]. In Salmonella typhimurium, the expression of class 1 genes (i.e., flhD and flhC) activates expression of genes required for flagella assembly and regulates expression class 2 genes (e.g., fliAZY and flhBAE), which in turn regulates expression of class 3 genes encoding flagellar structural proteins (e.g., fliC, flgMN, and MotAB) [18]. In Xenorhabdus nematophila, it was shown that the EnvZ-OmpR-FlhDC-FliA regulatory network coordinately controls flagella synthesis as well as exoenzyme and antibiotic production [8].
In this paper, we describe the transcriptional regulation of fliC and flhA expression by flhD/C and also show that flhD/C has an effect on extracellular secretion of the Carocin S1 protein, but not on Carocin S1 gene expression. Our results indicate that the type III secretion system of Pectobacterium carotovorum subsp. carotovorum has a new secretory function.
Methods
Bacterial strains, plasmids, media, and growth conditions
The strains and plasmids used are shown in Table 1. Pectobacterium carotovorum subsp. carotovorum strains were propagated at 28°C in 1.4% nutrient agar (NA) or with shaking in Luria-Bertani (LB) medium with NaCl (5 g/L). E. coli strains were propagated at 37°C in LB medium with shaking. Rifampicin, kanamycin, and ampicillin (all at 50 mg/L) were added to either medium when necessary.
Table 1.
Bacteria and plasmids used in this study
| Bacterium or plasmid | Relevant characteristics | Source |
|---|---|---|
| E. coli | ||
| 1830 | pro-met-KmrNmr, containing transposon Tn5 on the ?sucidal? plasmid pJB4JI | Gantotti et al. [37] |
| DH5 | supE44hsdR17recA1endA1gyrA1thi-1relA1 | Hanahan and Reusch et al [26,38] |
| Pectobacterium carotovorum subsp. carotovorum | ||
| 89-H-4 | putative biocontrol agent | Laboratory stock |
| H-rif-8-6 | 89-H-4, Rifr | this work |
| Ea1068 | wild type | Laboratory stock |
| T-29 | wild type | Laboratory stock |
| E108 | wild type | Laboratory stock |
| A-100 | wild type | Laboratory stock |
| 86-H-2 | wild type | Laboratory stock |
| TH12-2 | H-rif-8-2, flhC:: Tn5, Rifr, Kanr | this work |
| KH17 | H-rif-8-2, flh D::Kan, Rifr, Kanr | this work |
| FliC-KO | H-rif-8-2, fli C::Kam, Rifr, Kamr | this work |
| FlhA-KO | H-rif-8-2, flh A::Kam, Rifr, Kamr | this work |
| plasmid | ||
| pBR322 | Ampr, Kanr | Bolivar et al [39] |
| pBYL2DC | Ampr, flhDC | this work |
| pBYL2C | Ampr, flhC | this work |
| pBYL2D | Ampr, flhD | this work |
| pBFC | Ampr, fliC | this work |
| pBFA | Ampr, flhA | this work |
Ampr indicates ampcillin resistance, Rifr indicates rifampicin resistance, and Kanr indicates Kanamycin resistance.
Bacterial mating
Bacterial mating was carried out on NA using the membrane-filter mating method [14] with 0.22-μm pore size membrane filters (Millipore, Inc. Bedford, MA). The filters were placed on NA and incubated overnight at 28°C. Appropriate dilutions of each progeny suspension were spread on modified Drigalski's agar plates [19] containing 50 μg/ml rifampicin and kanamycin and incubated at 28°C for 24–48 h before the colonies were isolated.
Bacteriocin assays
Bacteriocin production was examined as described previously [20] in hard IFO-802 (with 1.4% agar) and soft IFO-802 (with 0.65% agar) medium. Growth inhibition zones around the colonies were considered as an indication of bacteriocin production.
Genetic engineering techniques
Previously described techniques were used to isolate the plasmids of Pectobacterium carotovorum subsp. carotovorum [21,22] and E. coli [23]. Total DNA was isolated as previously described [22]. Oligonucleotide DNA primers were synthesized by MDE Bio Inc. (Taipei, Taiwan). Reagents were purchased from Takara Co. (Tokyo, Japan). Previously detailed protocols were utilized for the general polymerase chain reaction (PCR) [24] and thermal asymmetric interlaced PCR (TAIL-PCR) [25], except that in the latter technique the annealing temperature of specific primers was decreased from 63°C to 60°C. For TAIL-PCR, specific primers complementary to the respective sequences of Tn5 (PR-1, PR-2, PR-3, PF-1, PF-2, and PF-3) or known sequences after the first TAIL-PCR analysis (TH12-2F1, TH12-2F2, TH12-2R1, and TH12-2R2) were synthesized (Table 2). In addition, three arbitrary degenerate primers designated N-1, N-2, and N-3 were used (Table 2).
Table 2.
Primers used in this studya
| Primer | Sequence (5'→3') | |
|---|---|---|
| PR-1 | ......... | 5'-GCCGAAGAGAACACAGATTTAGCCCA |
| PR-2 | ......... | 5'-CCGCACGATGAAGAGCAGAAGTT |
| PR-3 | ......... | 5'-CAGATCTCTGGAAAACGGGAAAGG |
| PF-1 | ......... | 5'-AGAGAACACAGATTTAGCCCAGTCGG |
| PF-2 | ......... | 5'-CCGCACGATGAAGAGCAGAAGTTAT |
| PF-3 | ......... | 5'-GATCCTGGAAAACGGGAAAGGTTC |
| TH12-2F1 | ......... | 5'-GATGGTGAAATTGGCAGAAAC |
| TH12-2F2 | ......... | 5'-GGACATTAGTCCGGTTTGTTG |
| TH12-2R1 | ......... | 5'-CAACAAACCGGACTAATGTCC |
| TH12-2R2 | ......... | 5'-GTTTCTGCCAATTTCACCATC |
| N-1 | ......... | 5'-NGTCGA(G/C)(A/T)GANA(A/T)GAA |
| N-2 | ......... | 5'-GTNCGA(C/G)(A/T)CANA(A/T)GTT |
| N-3 | ......... | 5'-(A/T)GTGNAG(A/T)ANCANAGA |
| P-3 | ......... | 5'-CTCGACGTTGTCACTGAAGCGGGAAG |
| P-4 | ......... | 5'-AAAGCACGAGGAAGCGGTCAGCCCAT |
| DY-SR1 | ......... | 5'-GAAATCGATCACCGCCTTCACAC |
| DY-SF1 | ......... | 5'-AAAGAATTCTTCAGTCGCGTTG |
| flhA-sen | ......... | 5'-TCACTCAACGTTGCATCTAC |
| flhA-anti | ......... | 5'-CAAGATGTTGGCCAACAGATG |
| fliC-sen | ......... | 5'-TCGGTGCGAATGATGGTG |
| fliC-anti | ......... | 5'-AACGCAGCAGTGACAGC |
| fliC-Fu-sen | ......... | 5'-TGGTTTTATCCACGACTCAC |
| fliC-Fu-anti | ......... | 5'-ATGCAGCAGGATCCAGAAC |
| flhA-Fu-sen | ......... | 5'-TCACTCAAGCTTGCATCTAC |
| flhA-Fu-anti | ......... | 5'-CGGATTGTCGACTAGCTGG |
a All primers were purchased from MDE Bio Inc., Taipei, Taiwan
TAIL-PCR products were sequenced using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA). Cycle sequencing was carried out in a GeneAmp System 9600 thermocycler (Applied Biosystems). Sequencing was carried out according to the manufacturer's protocol using an ABI 373S automated DNA sequencer 373S (Applied Biosystems).
Southern and colony hybridizations, probe labeling, and detection were performed by using a DIG DNA Labeling and Detection kit (Boehringer Mannheim GmbH, Mannheim, Germany) as described by the manufacturer. Hybridization was performed overnight, and the membrane was washed according to the recommendations of the manufacturer.
DNA electrophoresis, restriction digest, ligation, and transformation procedures for E. coli were performed as previously described [24]. Plasmid DNA transformation for Pectobacterium carotovorum subsp. carotovorum was performed using two previously described methods [26,27] following an incubation at 35°C until the optical density (550 nm) of the culture was 0.40 to 0.55.
Subcloning of flhD/C DNA from H-rif-8-6
The DNA fragment of flhD/C was amplified by PCR from H-rif-8-6 using oligonucleotide primers DY-SF1 and DY-SR1. The flhD/C DNA containing product was digested with restriction enzymes ClaI and EcoRI and subcloned into plasmid pBR322. The new plasmid was designated pBYL2DC. One hundred transformed colonies were isolated using selective LB agar containing 100 μg/ml of ampicillin after the transfer of pBYL2DC into E. coli DH05. The presence of the flhD/C DNA was detected by colony hybridization using flhD/C DNA probes and electrophoresis after digestion with ClaI and EcoRI to yield the expected 1.3-Kb DNA fragment bearing flhD/C. The pBYL2DC DNA was isolated from DH05/pBYL2DC and transferred into the insertion mutants of Pectobacterium carotovorum subsp. carotovorum TH12-2. One hundred colonies were isolated by selection on modified Drigalski's medium containing 50 μg/ml of kanamycin, rifampicin, and ampicillin. The flhD/C DNA was detected as previously described.
Construction of the null alleles of flhD, fliC, and flhA genes
The flhD gene was isolated from pBYL2DC by digesting with BsmI, which cleaves at two sites in pBYL2DC and thereby conveniently deletes flhC from the operon. The resulting plasmid was designated pBYL2D. A kanamycin resistant gene from pACYC177 was isolated, made blunt-ended using a DNA-blunting kit (Takara Co., Tokyo, Japan), and inserted in the unique EcoRV site of the flhD gene. The resulting plasmid was designated pBYL2D-Kan. The pBYL2D-Kan was re-isolated and linearized after HpaI and SspI restriction enzyme digestion, which deleted the ampicillin resistance gene and replication site of the plasmid. The linearized construct was transferred into H-rif-8-6, resulting in the homologous replacement of the native flhD gene and generating a null allele.
The DNA fragment of fliC was amplified by PCR from H-rif-8-6. After PCR amplification using two oligonucleotide primers (fliC-sen and fliC-anti), the partial fliC DNA fragment was purified, digested using AhdI and HindIII, and subcloned into plasmid pBR322 to generate the fliC plasmid. A kanamycin resistant gene from pACYC177 was isolated, made blunt-ended, and inserted into the unique SalI site of the fliC gene. The resulting plasmid was designated pfliC-Kan. The pfliC-Kan was linearized after AhdI and HindIII restriction enzyme digestion, which deleted the ampicillin resistance gene and replication site of the plasmid. The linearized construct was transferred into H-rif-8-6 resulting in the homologous replacement of the native fliC gene and generating a null allele.
The DNA fragment of flhA was amplified by PCR from H-rif-8-6 using oligonucleotide primers flhA-sen and flhA-anti. The partial flhA DNA fragment was purified, digested using the restriction enzymes ClaI and EcoRI, and subcloned into plasmid pBR322 using T4 ligase to generate the flhA plasmid. A kanamycin resistant gene from pACYC177 was isolated, made blunt-ended, and inserted in the unique SalI site of the flhA gene. The resulting plasmid was designated pflhA-Kan.
Computer analysis of sequence data
The nucleotide sequence and the deduced amino-acid sequence of FlhD/C were compared using the BLAST and FASTA programs of the National Center for Biotechnology Information server. Sequence data were compiled by DNASIS-Mac software (Hitachi, Tokyo, Japan).
RNA preparation and Northern hybridization
Bacteriocin synthesis medium (BSM; 0.5% sucrose, 0.1% NH4Cl, 0.2% KH2PO4, and 0.02% MgSO4·7H2O [pH = 7.5]) was used to produce Carocin S1. Total RNA was extracted from cells (Pectobacterium carotovorum subsp. carotovorum harboring constructs) that were grown without drugs at 28°C. To determine the stability of H-rif-8-6, TH12-2, TH12-2/pBYL2C, KH17, and KH17/pBYL2D strains, culture samples (8 ml each; with rifampicin [0.2 mg/ml] added when cell density was ~150 Klett units to block bacterial contamination) were withdrawn at various time points and transferred to tubes containing 5 mL of ice-cold water. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Northern blot hybridizations were performed using 10 μg of total RNA. RNA samples were denatured in RNA sample buffer at 65°C for 10 min. The buffer consisted of 250 μL formamide, 83 μL of 37% (w/v) formaldehyde, 83 μL of 6× loading dye (Promega, Madison, WI), 50 μL of 10× morpholinepropanesulfonic acid (MOPS; 20 mM MOPS and 5 mM sodium acetate) buffer, 1 mM EDTA (pH 7.0), and 34 μL of distilled water. RNA samples were separated on 1% agarose gels containing MOPS buffer with 2% (v/v) formaldehyde. DNA probes were synthesized by PCR using specific oligonucleotides (template sequences): PCAR-R3 (for caroS1K), PflhC-R1 (for flhC), and PflhD (for flhD) derived from Pectobacterium carotovorum subsp. carotovorum (Table 2). Template DNAs (caroS1K, flhD, and flhC) were obtained by PCR amplification. The probes were nonradioactively labeled by random priming using a digoxigenin (DIG) High Prime kit (Roche, Basel, Switzerland). To add the correct amount of probe for hybridization, a serial dilution of each probe (0.05–10 pg) was spotted on a nylon membrane, and the labeling sensitivity (amount of labeled DNA per spot) was determined. RNA was transferred overnight to a positively charged nylon membrane (Amersham Biosciences, Buckinghamshire, England) by capillary transfer using 20× SSC (0.3 M NaCl and 0.03 M sodium citrate, pH 7.0). The membrane after hybridization (performed for 16 h at 50°C in DIG Eazy Hyb buffer solution; Roche) was washed, and the specific transcripts on the blots were detected using a DIG luminescence detection kit (Roche) according to the manufacturer's protocol.
Motility test
A sterile loopful of bacterial cells was carefully inoculated vertically into tubes containing soft agar (IFO-802 medium with 0.5% agarose). After incubation for one month, motility was determined by migration and/or outgrowth of bacterial cells from the original inoculation line.
Results
Isolation of transposon insertion mutants
Conjugation of strain H-rif-8-6 with E. coli 1830 led to the isolation of 3000 colonies that grew on the selective plates containing 50 μg/mL rifampicin and kanamycin. Their antibiotic resistance was ascertained by rechecking growth on the selective medium and was found to be a stable property.
Bacteriocin assay of Tn5 insertional mutants
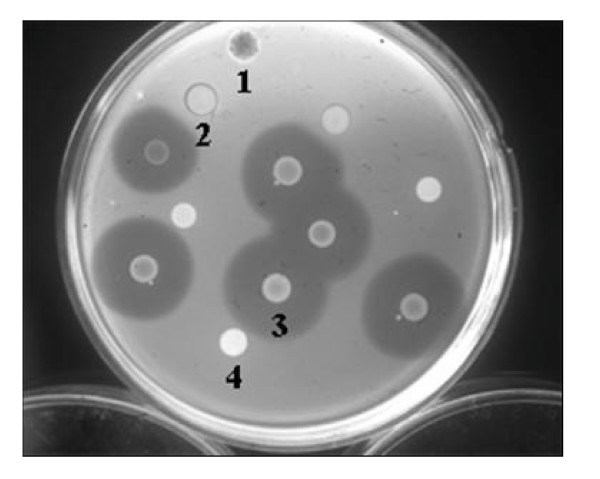
The bacteriocin activity of the putative insertion mutants was examined. The diameters of the inhibition zone typical were smaller around the putative mutant strains than parental strains, indicating the possibility that a gene related to Carocin S1 production had been inserted into the Tn5 transposon (Fig. 1).
Figure 1.
Bacteriocin activity of Tn5 insertion mutants of the Pectobacterium carotovorum subsp. carotovorum strains: 1, Serratia sp. (marker); 2, TH12-2 (Tn5 insertion mutant, flhC::Tn5); 3, H-rif-8-6 (parent); 4, E. coli 1830/pJB4JI (containing Tn5). The unlabeled strains are all Tn5 insertion mutants of the H-rif-8-6 parental strain. Strain Ea1068 was used as an indicator for bacteriocin activity.
Detection of Tn5 insertions in the mutants
To ascertain whether a Tn5 insertion had actually occurred in the putative mutant strains, nested-PCR was used to amplify the nptII gene [28] using the oligonucleotide primers P-3 and P-4 (Table 2). A total of 97% of the test isolates but not H-rif-8-6 produced a 500-bp DNA fragment that did not harbor the Tn5 insertion. Southern blot hybridization confirmed these results (data not shown).
Amplification of the DNA at the Tn5 insertion junction site and sequence analysis
TAIL-PCR was used to analyze the DNA sequences at the junctions of the Tn5 insertions. After the first TAIL-PCR experiment, two or more differently sized DNA fragments were obtained from each sample. All fragments were isolated by electrophoresis, purified, and sequenced and corresponding DNA fragments were shown to have the same sequence. Based on the sequence obtained from the first TAIL-PCR experiment, specific primers (TH12-2F1, TH12-2F2, TH12-2R1, and TH12-2R2) were synthesized for a second TAIL-PCR experiment. Subsequently, a nucleotide sequence of 1963 base pairs was obtained. The direction of transcription determined by analysis of the Tn5 insertions showed that two complete open reading frames (ORF2 and ORF3) were present and that Tn5 was located in ORF3 between base pairs 1312 and 1313. The 3' end of another open reading frame, ORF1, was located upstream of ORF2, and the 5' end of ORF4 was located downstream from ORF3 (Fig. 2).
Figure 2.
Nucleotide sequence of the flhD and flhC genes with the deduced amino-acid sequence of their respective proteins (FlhD and FlhC). The nucleotide sequence of fragments (positions 497-68 and 875-1453) represent flhD and flhC genes, respectively.
Homology with other genes and proteins
The predicted amino-acid sequences of ORF2 and ORF3 were compared to other known genes using the Swiss-Prot protein sequence data bank. A significant similarity was found between ORF2 and ORF3 of Pectobacterium carotovorum subsp. carotovorum and the flhD and flhC genes, respectively, of Pectobacterium carotovorum subsp. atroseptica (95% similarity), Serratia marcescens (86% similarity), Yersinia enterocolitica (84% similarity), and E. coli (80% similarity). Thus, ORF2 was designated as flhD, and ORF3 as flhC.
Bacteriocin expression, isolation, and activity assay
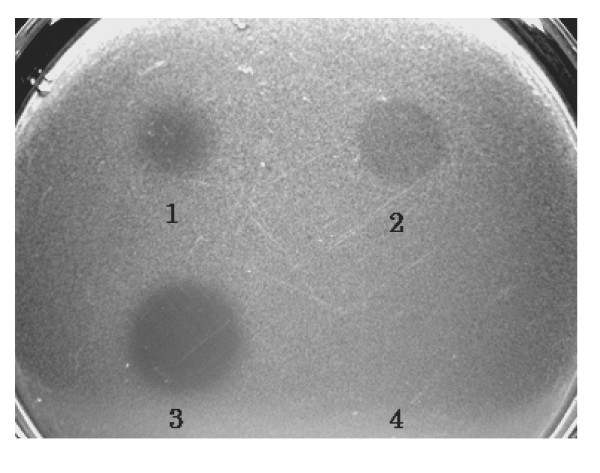
Bacteria in BSM medium were incubated in a sterilized stainless steel box with a stainless steel cover at 28°C for 24 h without any light. After centrifugation, the extracellular solution and cells were separated and collected. The cells were homogenized by sonication, and ammonium sulfate was added to 80% saturation to precipitate the protein. The precipitate was collected on a 0.45-μm cellulose filter. One milligram of precipitated protein was dissolved in 100 μl of bacteriocin buffer (0.1 M Tris [pH 7.5], 0.01 M DTT, and 0.5 M MgCl2). To determine bacteriocin antibiotic activity, 100 μg/10 μl of the CaroS1K protein solution was added to an indicator plate containing the Ea1068 strain growing on soft IFO-802 medium containing 0.65% agar. Growth inhibition zones at the point of addition were considered an indication of Carocin S1 activity (Fig. 3).
Figure 3.
Analysis of the killing activity of purified Carocin S1. Intracellular solution was isolated from Hi-rif-8-6 (1) and TH12-2 (3) strains. Extracellular solutions from Hi-rif-8-6 (2) and TH12-2 (4) strains were assayed for killing activity by addition to indicator plates containing strain Ea1068.
Isolation of null alleles of the flhD, fliC, and flhA genes
Since flagella assembly requires the expression of both the flhD and flhC genes, we constructed the strain FlhD-KO (flhD::Kan). The linearized construct (containing the flhD::Kan DNA fragment) was transferred into H-rif-8-6, resulting in the homologous replacement of the native flhD gene and generating a null allele. The resultant kan and rif resistant transformants were screened by PCR with one set of primers (DY-SR1 and DY-SF1) representing the 5' and the 3' termini of the flhD/C operon. This set of primers generated a 1.3-kb product, if the transforming DNA was not integrated. However, a homologous replacement of the native flhD gene by the null allele yielded a 2.7-kb product. The observed PCR product was 2.7 kb, indicating that the flhD gene had been replaced by the null allele. The gene was therefore designated as ΔflhD (strain KH17).
To confirm that Carocin S1 was actually secreted via T3bSS, we selected two components of T3bSS for deletion analysis, the fliC and flhA genes. The fliC gene encodes a FliC protein, which is an outer membrane component of T3bSS. The linearized construct (containing the fliC::Kan DNA fragment) was transferred into H-rif-8-6, resulting in the homologous replacement of the native fliC gene and generating a null allele. The kan and rif resistant transformants were screened by PCR with one set of primers (fliC-sen and fliC-anti) representing the 5' and the 3' termini of the fliC operon. The gene was therefore designated as ΔfliC (strain FliC-KO). The flagellin-associated gene flhA encodes the inner membrane FlhA component of T3bSS. The same procedure was used to obtain the flhA knockout (KO) mutant, and the gene was designated ΔflhA (strain FlhA-KO).
Complementation and analysis of flhD, flhC, fliC, and flhA genes
Wild-type H-rif-8-6 was used as a control and transformed with plasmids containing the flhD (pBYL2D) and flhC (pBYL2C) genes as well as the flhD/C (pBYL2DC) operon. The effect of these transformations on the bacteriocin production and cell size of the wild-type strain was assessed. The mutations in the flhC gene (TH12-2::Tn5) and the ΔflhD (KH17) gene were complemented by introduction of the flhD+(pBYL2D), flhC+(pBYL2C), and flhD/C (pBYL2DC) genes, and the effects of these respective genes were evaluated (see below).
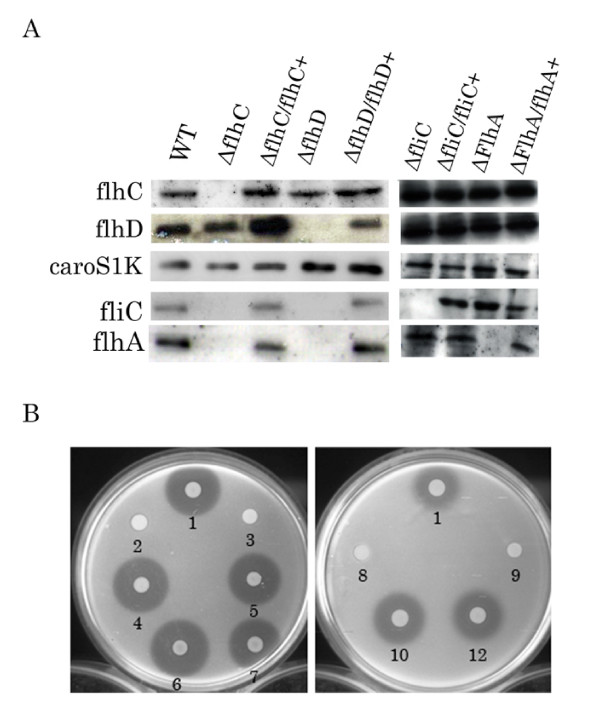
Plasmids pBYL2DC (containing flhD/C gene), pBYL2C (containing the flhC gene), pBYL2D (containing the flhD gene), pBFC (containing the fliC gene), and pBFA (containing the flhA gene) were expressed from their own native promoters in Pectobacterium carotovorum subsp. carotovorum TH12-2, KH17, FliC-KO, and FlhA-KO strains. Northern blot analysis of total RNA from H-rif-8-6, TH12-2, TH12-2/pBYL2C, KH17, KH17/pBYL2D, FliC-KO/pBFC, and FlhA-KO/pBFA cells incubated in BSM medium at 28°C for 24 h. PCR products specific for flhD, flhC, fliC, flhA, and caroS1K were used as probes in the hybridizations. The data indicate the presence of caroS1K gene in all strains and the presence of the other genes in all strains except the uncomplemented mutants, as expected (Fig. 4A).
Figure 4.
Bacteriocin activity and transcription analysis of Pectobacterium carotovorum subsp. carotovorum. (A) Transcription analysis of the flhD, flhC, caroS1K, and fliC genes. Total RNA (20 μg) from H-rif-8-6 (WT), TH12-2 (ΔflhC), TH12-2/pBYL2C (ΔflhC/flhC+), KH17 (ΔflhD), KH17/pBYL2D (ΔflhD/flhD+), FliC-KO (ΔfliC), FliC-KO/pBFC (ΔfliC/fliC+), FlhA-KO (ΔflhA), and FlhA-KO/pBFA (ΔflhA/flhA+) cells incubated in BSM medium at 28°C for 24 h was subjected to Northern blot analysis. Strain Ea1068 was used as an indicator for bacteriocin activity. (B) Bacteriocin activity assay. Numbered strains: 1, H-rif-8-6 (wild type); 2, TH12-2 (ΔflhC); 3, KH17 (ΔflhD); 4, TH12-2/pBYL2C (ΔflhC/flhC+); 5, TH12-2/pBYL2DC (ΔflhC/flhDC+); 6, KH17/pBYL2D (ΔflhD/flhD+); 7, KH17/pBYL2DC (ΔflhD/flhDC+); 8, FliC-KO (ΔfliC); 9, FlhA-KO (ΔflhA); 10, FliC-KO/pBFC (ΔfliC/fliC+); and 11, FlhA-KO/pBFA (ΔflhA/flhA+). Strain Ea1068 was used as an indicator for bacteriocin activity.
Bacteriocin activity assay for complementation
Assay of bacteriocin secreted from the insertion mutants, with and without complementation, indicated that, after complementation, mutants recovered the ability to secrete LMWB. Their larger inhibition zones were comparable in diameter to those of their parent strain, H-rif-8-6 (Fig. 4B). Neither the KH17 nor TH12-2 strains could secrete Carocin S1. However, complementation (by transformation of KH17 and TH12-2 with the flhD and flhC genes), respectively, rescued the ability of these strains to secrete Carocin S1 and thereby increased inhibition zone diameters, which were comparable in size to that of wild type. After transformation, all deletion strains harboring their respective complementing plasmids secreted LMWB (Fig. 4B).
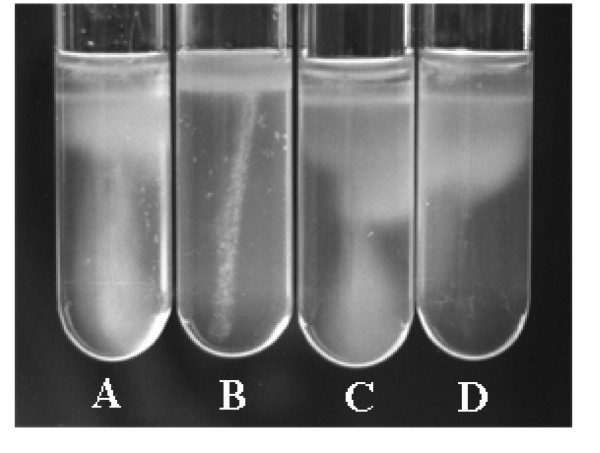
Motility
The wild-type strain, H-rif-8-6, but not the transposon insertion mutant, TH12-2, was motile (Fig. 5). The motility of TH12-2 was restored by transformation with the flhC (pBYL2C) and flhD/C (pBYL2DC) genes.
Figure 5.
Assay of motility in IFO-802 medium containing 0.5% agar, incubated at 25°C, over one month. A: H-rif-8-6 (parent), B: TH12-2 (ΔflhC), C: TH12-2/pBYL2DC (ΔflhC/flhDC+), and D: TH12-2/pBYL2C (ΔflhC/flhC+).
Discussion
In this study, the sequences of the flhD and flhC genes from Pectobacterium carotovorum subsp. carotovorum were highly homologous to the reported sequences of flhD/C genes in other bacterial strains [9-11,29,30]. These genes are adjacent and appear to share the same promoter [11]. Cloning of the flhD/C gene and subsequent transfer into the insertion mutant TH12-2 (flhC::Tn5) resulted in the recovery of bacteriocin activity (secretion of Carocin S1) in this mutant. The homologous replacement of the flhD gene by its null allele also resulted in the inhibition of Carocin S1 production. This indicated that both flhD and flhC are required for the production of Carocin S1 and, therefore, that the entire flhD/C operon influences the production of Carocin S1. FlhD has been previously shown to be associated with other stress-response systems [29,31]. Interestingly, flagella formation is controlled by the flhD/C operon [29]. In Gram-negative bacteria, the flagellar system is also known as the type III bacterium-flagella secretion system. Expression of the flhD/C genes is a form of response to environmental stress and requires the heat shock proteins DnaK, DnaJ, and GrpE [23], which are all related to environmental stress. Furthermore, the microcin B12 (mcbA) promoter is positively regulated by flhD [32,33]. It is therefore entirely appropriate to suggest that Carocin S1, which is normally induced by stress inducers like UV light and high competition from other related bacterial strains, is also under the control of flhD/C.
Although flhD/C was shown to control extracellular protein production through cumulative effects on hexA and gacA expression, this result was only demonstrated at the level of RNA transcription [34]. In this study, both the flhC and flhD genes regulated Carocin S1 secretion but not the transcription of the LMWB mRNA, caroS1K. Furthermore, assay of bacteriocin activity from TH12-2 (ΔflhC) detected intracellular but not extracellular Carocin S1 protein (Fig. 3). Similarly, we also found the transposon Tn5 insertion mutant, TH12-2 (ΔfliC), lost the ability to produce LMWB (data no shown). Northern blot analysis to monitor the expression of the caroS1K and fliC genes in the TH12-2 and KH17 strains detected the expression of caroS1K mRNA but not expression of fliC mRNA (Fig. 4A). However, as mentioned above, flhD/C genes regulate Gram-negative flagella synthesis and cell motility.
These results suggest that the flhD/C genes regulate the synthesis of bacterial flagella, which function as a flagellar type III secretion system (T3bSS) in Gram-negative bacteria, and that Carocin S1 utilizes this secretion machinery in Pectobacterium carotovorum subsp. carotovorum. However, because the growth of TH12-2 (fliC::Tn5) was extremely poor, this strain was lost before further experiments could be conducted. To further support our hypothesis, homologous replacement of the fliC gene (an outer membrane component of T3bSS) and the flhA gene (an inner membrane component of T3bSS) was used to generate a fliC null allele and flhA null allele, respectively. Bacteriocin analysis of extracellular fluids from the FliC-KO (fliC::kan) and FlhA-KO (flhA::Kan) strains also indicated significant inhibition of LMWB secretion. These results were similar to those found for TH12-2. Importantly, all these mutants still expressed the caroS1K mRNA.
The above results suggest a new function for the type III secretory system in this bacterial strain. Interestingly, complementation analysis of the fliC and flhA genes sometimes produced a smaller bacteriocin inhibition zone (3–8 mm versus 8 mm for the wild type). These results indicated that although the fliC and flhA genes are expressed in the FliC-KO/pBFC and FlhA-KO/pBFA strains, the secretion of the CaroS1K protein is not as efficient as in the wild-type strain, H-rif-8-6. In this study, the fliC and flhA genes were inserted into FliC-KO and FlhA-KO cells using multicopy plasmids for overexpression. It is therefore possible that the FliC or FlhA protein is not efficiently recruited into the T3bSS, and consequently CaroS1K cannot be secreted competently. Interestingly, the results of flhG [16] and fliC [15] gene complementation are similar to those found in our studies. These studies also support our hypothesis.
In previous studies, just one mechanism was utilized by Gram-negative plant and animal pathogens for T3bSS secretion and translocation of virulence determinants into susceptible eukaryotic cells [17]. The present study uniquely demonstrates that Pectobacterium cells can transfer Carocin S1 extracellularly using the T3bSS system and kill related bacterial cells.
The observed smaller size of flhD mutant cells confirms the observation of Prüss and Matsumura [35-39] and corroborates the suggestion that flhD is responsible for cell elongation. Interestingly, TH12-2 (flhC::Tn5) cells are longer (our unpublished data), which indicates that flhC also controls cell elongation. This is similar to what was observed in brg insertion mutants [6], indicating a possible interference with or disruption of cell division. This is directly opposite to what was observed in flhD mutants. It could therefore be proposed that though flhD inhibits cell division [31,35], flhC may promote cell division in this bacterial strain. Therefore, the flhC gene may have functions unrelated to its role in the flagellar regulon, which may be opposite to that of flhD. However, both flhD and flhC are required for determining bacterial cell size.
Conclusion
Based on these results, we conclude that the extracellular export of LMWB, Carocin S1, by Pectobacterium carotovorum subsp. carotovorum utilizes the type III secretion system, which also controls this bacterium's cell motility and cell size.
Authors' contributions
YC participated in the bacteriocin analysis and construction of the null alleles of the fliC and flhA genes. DC conceived the study, participated in its design, and corrected the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yung-chieh Chan, Email: d9451104@mail.nchu.edu.tw.
Huang-Pin Wu, Email: whanpyng@cgmh.org.tw.
Duen-yau Chuang, Email: chuang@dragon.nchu.edu.tw.
Acknowledgements
The support of this work by grants from the National Science Council (grants NSC-97-2313-B-005-027-MY3) of Taiwan (R.O.C.) is gratefully acknowledged.
References
- Chuang D-Y, Kyeremeh AG, Gunji Y, Takahara Y, Ehara Y, Kikumoto T. Identification and Cloning of an Erwinia carotovora subsp. carotovora bacteriocin regulator gene by insertional mutagenesis. J Bacteriol. 1999;181:1953–1957. doi: 10.1128/jb.181.6.1953-1957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaka K, Amako K. Morphology of bacteriocins. Pro Nuc Enzy. 1979;24:719–726. (in Japanese) [PubMed] [Google Scholar]
- Kikumoto T, Ma S, Takahara Y. Biological control of the soft rot disease of Chinese cabbage 3. Interactions of avirulent and virulent strains of Erwinia carotovora subsp. carotovora on the petiole of Chinese cabbage, abstr. 195. Abstracts of the papers presented at the Annual Meeting of the Society, 1993. The Phytopathological Society of Japan, Japan. 1993. pp. 315–316. (in Japanese)
- Kikumoto T, Kyeremeh AG, Chuang D-Y, Gunji Y. In: Proceedings of the Fourth International Workshop on Plant Growth-Promoting Rhizobacteria Japan-OECD Joint Workshop, Sapporo, Japan. Ogoshi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S, editor. 1997. Biological control of the soft rot disease of Chinese cabbage with avirulent mutant strains of Erwinia carotovora subsp. carotovora; pp. 118–119. [Google Scholar]
- Takahara Y. Development of the microbial pesticide for soft-rot disease. PSJ Biocont Rept. 1994;4:1–7. (in Japanese) [Google Scholar]
- Chuang D-Y, Gunji Y, Kyeremeh AG, Takahara Y, Kikumoto T. Cloning of bacteriocin regulator gene (brg) from Erwinia carotovora subsp. carotovora, abstr. 251. Abstracts of the papers presented at the Annual Meeting of the Society, 1998. The Phytopathological Society of Japan, Japan. 1998. pp. 389–390. (in Japanese)
- Chuang D-Y, Chien Y-C, Wu H-P. Cloning and Expression of the Erwinia carotovora subsp. Carotovora Gene Encoding the Low-Molecular- Weught Bacteriocin Carocin S1. J Bacteriol. 2007;189:620–626. doi: 10.1128/JB.01090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Forst S. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol Microbiol. 2006;61:1397–1412. doi: 10.1111/j.1365-2958.2006.05320.x. [DOI] [PubMed] [Google Scholar]
- Eberl L, Christiansen G, Molin S, Givskov M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J Bacteriol. 1996;178:554–559. doi: 10.1128/jb.178.2.554-559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen KL, Hughes KT. Negative regulatory loci coupling flagellin synthesis to flagellar assemby in Salmonella typhimurium. J Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givskov M, Ebert L, Christiansen G, Benedik MJ, Molin S. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- Galán JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- Devaux M, Hébraud M, Henderson IR, Pallen M. Type III secretion: what's in a name? Trends Microbiol. 2006;14:157–160. doi: 10.1016/j.tim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Gnerer J, Karlinsey JE, Hughes KT. Transcriptional and Translational Control of the Salmonella fliC Gene. J Bacteriol. 2006;188:4487–4496. doi: 10.1128/JB.00094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mizuno S, Aizawa S, Hughes KT. Mutations in Flk, FlgG, FlhA, and FlhE That Affect the Flagellar Type III Secretion Specificity Switch in Salmonella enteric. J Bacteriol. 2009;191:3938–3949. doi: 10.1128/JB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plant. Microbiol. Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey J, Pease AJ, Winkler ME, Bailey JL, Hughes KT. The flk Gene of Salmonella typhimurium Couples Flagellar P- and L-Ring Assembly to Flagellar Morphogenesis. J Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama H, Sakamoto M. Isolation methods of the soft-rot causing bacteria from the soil. Sci Rep Res Inst Tohoku Univ. 1952;3:29–34. [Google Scholar]
- Fredericq P. Colicins. Annu Rev Microbiol. 1957;11:7–22. doi: 10.1146/annurev.mi.11.100157.000255. [DOI] [PubMed] [Google Scholar]
- Kado CI, Liu ST. Rapid procedure for detection and isolation of the large and small plasmid. J Bacteriol. 1981;116:125–159. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita K. Experimental methods in soil microbiology. Soil Microbiological Society of Japan. 1. Youkendou Publishing Co., Japan; 1992. DNA Probes; pp. 163–172. (in Japanese) [Google Scholar]
- Shi W, Zhou Y, Wild J, Adler J, Gross CA. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacterial. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. 2. Cold Spring Laboratory press, Cold Spring Harbor, NY; 1989. [Google Scholar]
- Liu YG, Whittier RF. Thermal Asymmetric Interlaced PCR: Automatable Amplification and Sequencing of Insert End Fragments from P1 and YAC clones for Chromosome Walking. Genomic. 1994;25:674–681. doi: 10.1016/0888-7543(95)80010-J. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hinton JC, Perombelon MC, Salmond GP. Efficient Transformation of Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica. J Bacteriol. 1985;161:786–788. doi: 10.1128/jb.161.2.786-788.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima S, Hasebe A, Komoto Y, Charter JP, Miyashita K, Yokoyama K, Pickup RW. Detection of genetically engineered microorganisms in paddy soil using a simple and rapid "nested" polymerase chain reaction method. Soil Biol Biochem. 1995;27:219–227. doi: 10.1016/0038-0717(94)00162-T. [DOI] [Google Scholar]
- Dufour A, Furness RB, Hughes C. Novel genes that upregulate the Proteus mirabilis flhD/C master operon controlling flagellar biogenesis and swarming. Mol Microbiol. 1998;29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness RB, Fraser GM, Hay NA, Hughes C. Negative feedback from a Proteus class II flagellum export defect to the flhD/C master operon controlling cell division and flagellum assembly. J Bacteriol. 1997;179:5585–5588. doi: 10.1128/jb.179.17.5585-5588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell ND. PhD thesis. Harvard University, Cambridge, Mass; 1989. Regulation of a stationary phase promoter, Pmcb, in Escherichia coli. [Google Scholar]
- Tentler S. MS thesis. University of Illinois, Chicago; 1994. Gene regulation within the flhB operon of Escherichia coli. [Google Scholar]
- Cui Y, Chatterjee A, Yang H, Chatterjee K. Regulatory Network Controlling Extracellular Proteins in Erwinia carotovora subsp. carotovora: FlhDC, the Master Regulator of Flagellar Genes, Activates rsmB Regulatory RNA Production by Affecting gacA and hexA (lrhA) Expression. J Bacteriol. 2008;190:4610–4623. doi: 10.1128/JB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüss BM, Matsumura P. A regulator of the flagellar of Escherichia coli, flhD, also affects cell division. J Bacteriol. 1996;178:668–674. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüss BM, Matsumura P. Cell cycle regulation of flagellar genes. J Bacteriol. 1997;179:5602–5604. doi: 10.1128/jb.179.17.5602-5604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantotti BV, Kindle KL, Beer SV. Transfer of the drug-resistance transposon Tn5 to Erwinia herbicola and the induction of insertion Mutations. Curr Microbiol. 1981;6:417–425. doi: 10.1007/BF01567015. [DOI] [Google Scholar]
- Reusch RN, Hiske TW, Sadoff HL. Poly-beta-hydroybutyrate membrane structure and its relationship to genetic transformability in Escherichia coli. J Bacteriol. 1986;168:553–562. doi: 10.1128/jb.168.2.553-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Construction and characterization on of new cloning vehicles II. A multipurpose cloning system. Gene. 1977;2:95–113. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]