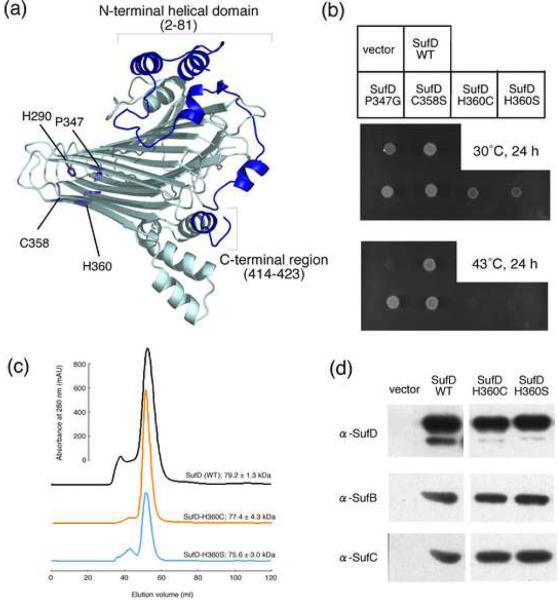
Fig. 4. Mutational Analysis of the SufD protein.
(a) The representation of a SufD subunit with its dimer interface in the front side. The amino acid residues changed by site-directed mutagenesis are depicted in the structure by stick representations. (b) Phenotypic characterization of the SufD mutations. Middle panel shows growth of the mutant (UT109) cells at the permissive temperature (30°C) of the temperature-sensitive plasmid, pKO3-NIF. Complementation for the loss of sufD is shown in the lower panel by the growth at 43°C. (c) Gel-filtration analysis of the mutant SufD proteins. The SufD proteins carrying N-terminally fused His-tag and the H360C or H360S mutation were expressed in the mutant E. coli cells (YT2512) in which the chromosomal sufABCDSE operon was deleted. The proteins were purified with Ni resin and then subjected to the gel-filtration (Sephacryl S-200) chromatography. The molecular sizes were estimated from the three experiments, and the values are the mean ± SD. (d) Pull-down assays to examine the interactions of the SufD mutant proteins with SufB and SufC. The SufD proteins carrying N-terminally fused His-tag and the H360C or H360S mutation were co-expressed in the YT2512 cells with intact SufB and SufC proteins using the plasmid pRK-sufABC-SE (ΔsufD). The proteins were purified with Ni resin and subjected to Western blot analysis using specific antibodies. Detection was with an ECL Plus kit (GE Healthcare).