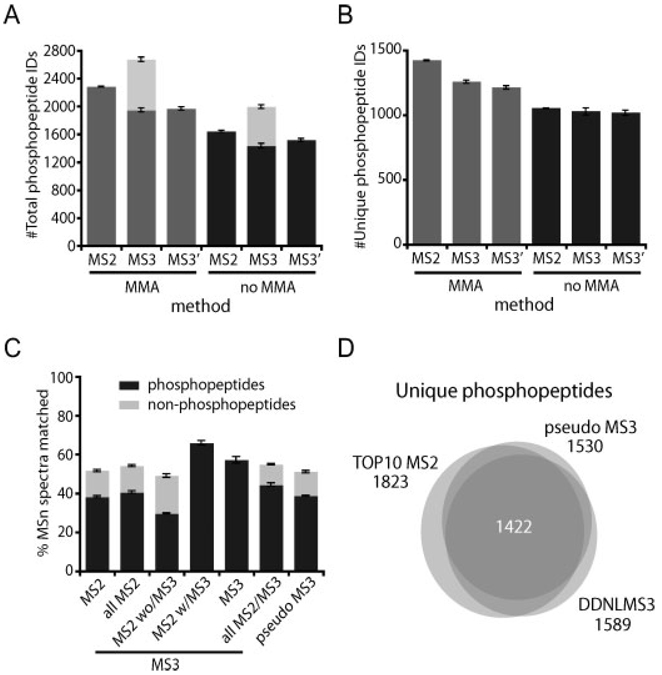
Figure 5.
Phosphopeptide identifications per analysis at <1% FDR using each of the three methods. Mean ± SD values for triplicate analyses are shown. (MS2 = TOP10 MS2 method, MS3 = DDNLMS3 method, MS3′ = pseudo MS3 method, MMA: searches at 50 ppm precursor mass tolerance, using mass accuracy information as a filtering criteria, no MMA: searches at 2.0 Da precursor mass tolerance, not using mass tolerance as a filter). (A) Total (redundant) phosphopeptide spectral matches. (B) Unique (non-redundant) phosphopeptide sequences. In order to be conservative, unique phosphopeptides were calculated at sequence level without considering different phosphorylation site localizations. (C) Success rate in phosphopeptide and unmodified peptide identification for each method. Values are calculated based on the total number of MSn scans collected. Details for different scan types are given in the MS3 method. (D) Overlap in phosphopeptide identification between the three methods. For simplicity, overlaps were calculated at sequence level without considering different phosphorylation site localizations. Each circle represents the combined triplicate analyses for each of the methods. A total of 2041 non-redundant phosphopeptides were identified in the SCX fraction #5 used in this study.