Abstract
The DNA-repair machinery is faced with the significant challenge of differentiating DNA lesions from unmodified DNA. Two recent publications, one in this issue of Nature Structural & Molecular Biology, uncover a new way of recognizing minimally distorting DNA lesions: insertion of a 3- or 4-amino-acid wedge into DNA to extrude the lesion into a shallow binding pocket that can accommodate various damaged bases.
Damaged bases are most frequently removed from DNA by one of two pathways: base-excision repair (BER) and nucleotide-excision repair (NER)1. In BER, small modifications to the DNA bases caused by oxidation, deamination or alkylation are recognized and excised by DNA glycosylases2. At least 12 such enzymes are known in humans; they show narrow substrate specificity and recognize one or a few prominent endogenous lesions. DNA glycosylases hydrolyze the glycosidic bond linking the base to the sugar-phosphate backbone, generating an abasic-site product, which is processed by a common set of downstream enzymes that remove the abasic site and restore the original DNA sequence. These enzymes recognize and extrude the damaged nucleotide from the DNA helix into an active site pocket that confers specificity for the damaged base. A single aromatic or aliphatic residue is inserted into the helix from the minor groove and takes up the position of the displaced base3,4.
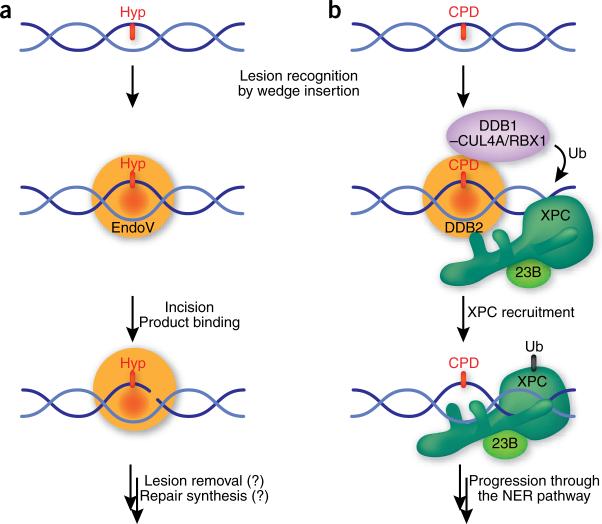
A few repair enzymes use slightly different strategies to recognize small base lesions. Among them is Endonuclease V (EndoV), which was originally isolated from Escherichia coli as an enzyme to process hypoxanthine arising from the nitrosative deamination of adenine5. EndoV also binds other deaminated bases, as well as mismatches, flap structures and Y structures, and it remains to be elucidated which one of these is the principal substrate in mammals6. Upon binding its substrate, EndoV cleaves the phosphodiester bond located one nucleotide 3′ to the lesion (Fig. 1a). This unusual mechanism for dealing with base lesions raises the question of whether EndoV also uses a nucleotide-flipping mechanism to recognize lesions in DNA.
Figure 1.
Recognition and repair pathway initiation of slightly distorting DNA lesions. (a) EndoV recognizes deaminated bases, in particular hypoxanthine (Hyp), by inserting a wedge into the DNA helix and binding the lesion in a specific pocket. The catalytic activity of EndoV makes an incision in the phosphodiester bond located one nucleotide 3′ to the lesion. EndoV remains bound to the incised product, allowing the recruitment of hitherto unknown downstream factors and completion of the pathway without exposure of the nick. (b) DDB2 binds to CPD (and other) lesions by inserting a wedge into the DNA helix to extrude the modified base into a shallow binding pocket. Binding of the DDB2–DDB1 complex to DNA locates the CUL4A/RBX1 ubiquitin ligase in proximity of the lesion, leading to the ubiquitination (Ub) of XPC and DDB2. This modification weakens the DNA binding activity of DDB2, but not that of XPC, and allows XPC–RAD23B (23B) to replace DDB2–DDB1 on the damaged site and progression through the NER pathway.
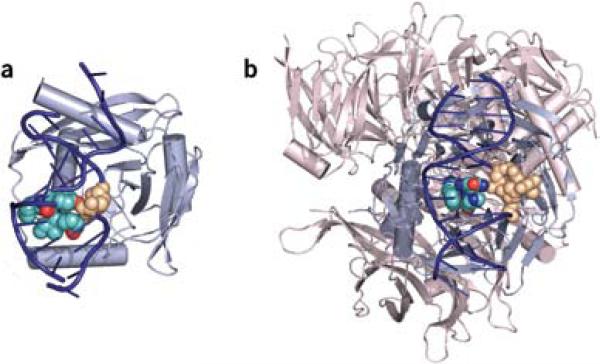
A paper by Dalhus et al.7 on page 138 of this issue of Nature Structure & Molecular Biology provides fascinating insights into this question. The distinguishing feature of the EndoV–DNA complex is a wedge of four residues (PYIP) that inserts into the minor groove of the helix (Fig. 2). In contrast to the precise extrusion of a single nucleotide by DNA glycosylases of almost 180° (ref. 3), the lesioned base in EndoV is rotated by only 90° into the binding pocket of EndoV, located on the minor groove side. The orphaned cytosine base in turn is slightly pushed into the minor groove of the helix. The role of the wedge might be to tap on the helix and present potentially modified bases to the lesion binding pocket. The protein stays bound to the 3′ incised product through a series of hydrogen bonds to the terminal 5′ phosphate at the excision position, with the hypoxanthine still fully engaged in the recognition pocket. This product binding probably serves to ensure that the nicked product is securely passed to the subsequent (currently unknown) enzymes in the pathway that remove the damage and restore the original DNA sequence.
Figure 2.
EndoV and DDB1–DDB2 complexes. (a) EndoV binds hypoxanthine by inserting a wedge made up of the amino acids PYIP (turquoise and red, CPK representation) from the minor groove of the helix and displacing the base (tan, CPK) into a shallow binding pocket located in the minor grove. DNA is shown in dark blue and EndoV in light blue. (b) UV-DDB binds to 6−4PP through the DDB2 subunit. DDB2 uses a wedge made up of the amino acids PNH (turquoise, blue and red, CPK) that binds from the minor groove of DNA and extrudes the UV-induced adduct (tan, CPK) into a shallow binding pocket in the major groove. DDB2 is shown in light blue; DDB1 is shown in light red and binds DDB2 on the opposite side of the DNA binding surface. DNA is dark blue. Figures were made using PyMOL (http://pymol.sourceforge.net) using the structures PDB 2W35 and PDB 2W36 for EndoV and PDB 3EI1 for UV-DDB.
So is this wedge mechanism for base extrusion and recognition found in other DNA-damage binding proteins? In a parallel and almost simultaneous study of a different repair pathway, NER, Scrima et al. report that UV DNA-damaged binding complex (UV-DDB, made up of the DDB1 and DDB2 subunits) uses a similar wedge-base mechanism to find lesions8. NER deals with numerous structurally diverse lesions formed by environmental agents such as solar UV radiation and others9. Within NER, UV-DDB is required for lesions that only moderately destabilize the DNA helix, whereas more distorting lesions are directly recognized by the XPC–RAD23B complex. XPC-RAD23B acts downstream of UV-DDB and recognizes the thermodynamic destabilization induced in the DNA by many NER substrates10 (Fig. 1b). UV-DDB is of particular importance for the repair of cyclopyrimidine dimers (CPD), which have only a mild influence on the structure of the DNA. The recent structure of DDB1–DDB2 bound to another photolesion, the 6−4 photoproduct (6−4PP), provides important insight into the mechanism of damage recognition in NER8. DNA is exclusively bound by the DDB2 subunit of the complex. DDB2 has a WD-40 β-propeller structure, and DNA binding is mediated by loops emerging from one side of the propeller. The key element for DDB2-mediated damage sensing is a 3-residue hairpin made up of the strictly conserved residues Phe371, Gln372 and His373 that inserts into the DNA from the minor groove, displacing the lesion into a shallow binding pocket located in the major groove (Fig. 2b). The wedge has striking geometric similarity to the one found in EndoV, and in both cases 2 amino acids are stacked at the tip of the wedge (Gln372 and His373 in DDB2 and Tyr80 and Pro82 in EndoV) and inserted into the DNA helix. As is seen in EndoV, the lesion binding pocket in DDB2 is shallow. It does not provide much specificity for the 6−4PP, but seems to be well suited to accommodate various lesions with an intrastrand cross-link, including CPDs and 6−4PPs. Cross-linked bases induce a compression of the phosphate backbone of the DNA, and this compression is readily accommodated in DDB2. These properties allow DDB2 to bind minimally distorting lesions.
The interaction mode of DDB2 with damaged DNA ideally complements the binding of XPC–RAD23. The binding of the DDB2 opens the DNA at the lesion and induces a kink, making the CPD recognizable for XPC, which interacts with the adducted site by binding 2 nucleotides of undamaged single-stranded DNA opposite the lesion11. UV-DDB is associated with the ubiquitin ligase CUL4A/RBX1 via the DDB1 subunit12. DDB2 binding to DNA anchors this complex at the damaged sites and this leads to ubiquitination of DDB2 itself and XPC at the site of the lesion13 (Fig. 1b). This event is thought to weaken the binding of DDB2 to the lesion, permitting XPC to displace DDB2 and recruit the core NER factors. Inspection of the DDB2 and XPC DNA structures suggests that a clash of the respective damage binding regions would not permit the two proteins to bind simultaneously, providing one possible reason for the existence of the ubiquitination cascade. The complementary binding strategies of DDB2 and XPC thus provide a powerful way of recognizing a broad variety of lesions that induce diverse structural alterations in the DNA helix. Once XPC binding has been established, downstream NER factors are recruited. These factors ensure that a chemical modification is indeed present in the DNA and that the distortion is not induced by a simple mismatch. They then excise the lesion, as part of an oligonucleotide of 24−32 nucleotides, and fill in the resulting DNA gap9.
Considering the terrific progress made in the structural biology of DNA-damage recognition by proteins in the past 15 years, it is remarkable that studies of two unrelated proteins reveal a novel mechanism for locating damaged sites in DNA, a wedge consisting of 3 or 4 amino acids. It will be interesting to see whether this structural element is also present in other proteins. At least one other protein, the ultraviolet damage endonuclease (UVDE), for which only a DNA-free structure is presently available, seems to contain a similar element14. All three proteins recognize a diversity of lesions, so the wedge may be an important feature in proteins with broad substrate recognition.
ACKNOWLEDGMENT
Work in the O.D.S. laboratory is supported by US National Institutes of Health grants GM080454 and CA092584. A.J.C. is supported by an Integrative Graduate Education and Research Trainseeship predoctoral fellowship (National Science Foundation Award No. 0549370).
Contributor Information
Orlando D Schärer, Departments of Pharmacological Sciences and Chemistry, Graduate Chemistry Building Room 619, Stony Brook University, Stony Brook, New York 11794−3400, USA. e-mail: orlando@pharm.stonybrook.edu.
Arthur J Campbell, Department of Chemistry, Graduate Chemistry Building Room 619, Stony Brook University, Stony Brook, New York 11794−3400, USA..
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. ASM; Washington DC: 2005. [Google Scholar]
- 2.Hegde ML, Hazra TK, Mitra S. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hitomi K, Iwai S, Tainer JA. DNA Repair (Amst.) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Yang W. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 5.Yao M, Hatahet Z, Melamede RJ, Kow YW. J. Biol. Chem. 1994;269:16260–16268. [PubMed] [Google Scholar]
- 6.Moe A, et al. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalhus B, et al. Nat. Struct. Mol. Biol. 2009;16:138–143. doi: 10.1038/nsmb.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scrima A, et al. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillet LC, Schärer OD. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 10.Sugasawa K, et al. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min JH, Pavletich NP. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 12.Groisman R, et al. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 13.Sugasawa K, et al. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 14.Paspaleva K, et al. Structure. 2007;15:1316–1324. doi: 10.1016/j.str.2007.05.010. [DOI] [PubMed] [Google Scholar]