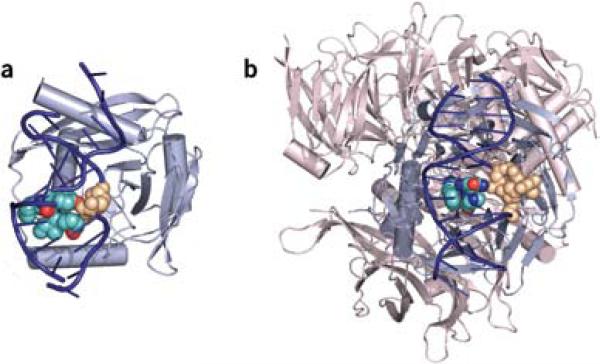
Figure 2.
EndoV and DDB1–DDB2 complexes. (a) EndoV binds hypoxanthine by inserting a wedge made up of the amino acids PYIP (turquoise and red, CPK representation) from the minor groove of the helix and displacing the base (tan, CPK) into a shallow binding pocket located in the minor grove. DNA is shown in dark blue and EndoV in light blue. (b) UV-DDB binds to 6−4PP through the DDB2 subunit. DDB2 uses a wedge made up of the amino acids PNH (turquoise, blue and red, CPK) that binds from the minor groove of DNA and extrudes the UV-induced adduct (tan, CPK) into a shallow binding pocket in the major groove. DDB2 is shown in light blue; DDB1 is shown in light red and binds DDB2 on the opposite side of the DNA binding surface. DNA is dark blue. Figures were made using PyMOL (http://pymol.sourceforge.net) using the structures PDB 2W35 and PDB 2W36 for EndoV and PDB 3EI1 for UV-DDB.